4EO5
 
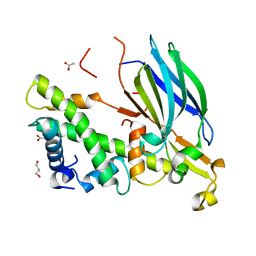 | | Yeast Asf1 bound to H3/H4G94P mutant | | 分子名称: | ACETATE ION, GLYCEROL, Histone H3.2, ... | | 著者 | Scorgie, J.K, Churchill, M.E. | | 登録日 | 2012-04-13 | | 公開日 | 2012-06-13 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | The conformational flexibility of the C-terminus of histone H4 promotes histone octamer and nucleosome stability and yeast viability.
Epigenetics Chromatin, 5, 2012
|
|
3KN1
 
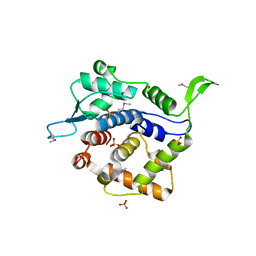 | | Crystal Structure of Golgi Phosphoprotein 3 N-term Truncation Variant | | 分子名称: | Golgi phosphoprotein 3, SULFATE ION | | 著者 | Schmitz, K.R, Bessman, N.J, Setty, T.G, Ferguson, K.M. | | 登録日 | 2009-11-11 | | 公開日 | 2009-12-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | PtdIns4P recognition by Vps74/GOLPH3 links PtdIns 4-kinase signaling to retrograde Golgi trafficking.
J.Cell Biol., 187, 2009
|
|
1M7S
 
 | | Crystal Structure Analysis of Catalase CatF of Pseudomonas syringae | | 分子名称: | Catalase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Carpena, X, Soriano, M, Klotz, M.G, Duckworth, H.W, Donald, L.J, Melik-Adamyan, W, Fita, I, Loewen, P.C. | | 登録日 | 2002-07-22 | | 公開日 | 2002-08-28 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of the Clade 1 catalase, CatF of Pseudomonas syringae, at 1.8 A resolution
Proteins, 50, 2003
|
|
1ME1
 
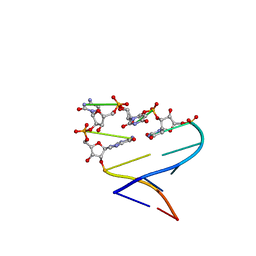 | | Chimeric hairpin with 2',5'-linked RNA loop and RNA stem | | 分子名称: | 5'-R(*GP*GP*AP*CP*(U25)P*(U25)P*(C25)P*(5GP)P*GP*UP*CP*C)-3' | | 著者 | Denisov, A.Y, Hannoush, R.N, Gehring, K, Damha, M.J. | | 登録日 | 2002-08-07 | | 公開日 | 2003-08-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A Novel RNA Motif Based on the Structure of Unusually Stable 2',5'-Linked r(UUCG) Loops
J.AM.CHEM.SOC., 125, 2003
|
|
4ERP
 
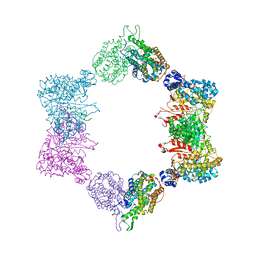 | |
4EPP
 
 | |
4EPQ
 
 | |
4ERM
 
 | |
1U0J
 
 | | Crystal Structure of AAV2 Rep40-ADP complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA replication protein | | 著者 | James, J.A, Aggarwal, A.K, Linden, R.M, Escalante, C.R. | | 登録日 | 2004-07-13 | | 公開日 | 2004-08-24 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of adeno-associated virus type 2 Rep40-ADP complex: Insight into
nucleotide recognition and catalysis by superfamily 3 helicases
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
8HN7
 
 | | Crystal structure of monoclonal antibody complexed with SARS-CoV-2 RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of monoclonal antibody 3C11, Light chain of monoclonal antibody 3C11, ... | | 著者 | Qi, J, Chen, Y. | | 登録日 | 2022-12-07 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-06-07 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Characterization of RBD-specific cross-neutralizing antibodies responses against SARS-CoV-2 variants from COVID-19 convalescents.
Front Immunol, 14, 2023
|
|
1YQA
 
 | |
1TPT
 
 | | THREE-DIMENSIONAL STRUCTURE OF THYMIDINE PHOSPHORYLASE FROM ESCHERICHIA COLI AT 2.8 ANGSTROMS RESOLUTION | | 分子名称: | SULFATE ION, THYMIDINE PHOSPHORYLASE, THYMINE | | 著者 | Walter, M.R, Cook, W.J, Cole, L.B, Short, S.A, Koszalka, G.W, Krenitsky, T.A, Ealick, S.E. | | 登録日 | 1990-06-14 | | 公開日 | 1991-07-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Three-dimensional structure of thymidine phosphorylase from Escherichia coli at 2.8 A resolution.
J.Biol.Chem., 265, 1990
|
|
1Z5C
 
 | |
1Q14
 
 | | Structure and autoregulation of the yeast Hst2 homolog of Sir2 | | 分子名称: | CHLORIDE ION, HST2 protein, ZINC ION | | 著者 | Zhao, K, Chai, X, Clements, A, Marmorstein, R. | | 登録日 | 2003-07-18 | | 公開日 | 2003-09-30 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure and autoregulation of the Yeast Hst2 homolog of Sir2
Nat.Struct.Biol., 10, 2003
|
|
1Z59
 
 | |
1RNB
 
 | |
1V4A
 
 | | Structure of the N-terminal Domain of Escherichia coli Glutamine Synthetase adenylyltransferase | | 分子名称: | Glutamate-ammonia-ligase adenylyltransferase | | 著者 | Xu, Y, Zhang, R, Joachimiak, A, Carr, P.D, Ollis, D.L, Vasudevan, S.G. | | 登録日 | 2003-11-12 | | 公開日 | 2004-07-27 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of the n-terminal domain of Escherichia coli glutamine synthetase adenylyltransferase
Structure, 12, 2004
|
|
3QBZ
 
 | | Crystal structure of the Rad53-recognition domain of Saccharomyces cerevisiae Dbf4 | | 分子名称: | DDK kinase regulatory subunit DBF4, SULFATE ION | | 著者 | Matthews, L.A, Jones, D.R, Prasad, A.A, Duncker, B.P, Guarne, A. | | 登録日 | 2011-01-14 | | 公開日 | 2011-12-07 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.692 Å) | | 主引用文献 | Saccharomyces cerevisiae Dbf4 has unique fold necessary for interaction with Rad53 kinase.
J.Biol.Chem., 287, 2012
|
|
1QL2
 
 | |
1Z5B
 
 | |
1T2R
 
 | | Structural basis for 3' end recognition of nucleic acids by the Drosophila Argonaute 2 PAZ domain | | 分子名称: | 5'-R(*CP*UP*CP*AP*C)-3', Argonaute 2 | | 著者 | Lingel, A, Simon, B, Izaurralde, E, Sattler, M. | | 登録日 | 2004-04-22 | | 公開日 | 2004-06-01 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Nucleic acid 3'-end recognition by the Argonaute2 PAZ domain.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1Z5A
 
 | | Topoisomerase VI-B, ADP-bound dimer form | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Type II DNA topoisomerase VI subunit B | | 著者 | Corbett, K.D, Berger, J.M. | | 登録日 | 2005-03-17 | | 公開日 | 2005-06-14 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural dissection of ATP turnover in the prototypical GHL ATPase TopoVI.
Structure, 13, 2005
|
|
1QGT
 
 | |
1QL1
 
 | |
3TD7
 
 | |
