5TB8
 
 | |
3CCU
 
 | |
3CC2
 
 | |
3CCL
 
 | |
3CCV
 
 | |
3CC7
 
 | |
3CCE
 
 | |
3CC4
 
 | |
3CCS
 
 | |
3CCR
 
 | |
3CCM
 
 | |
3CCJ
 
 | |
3CME
 
 | |
4EYV
 
 | | Crystal structure of Cyclophilin A like protein from Piriformospora indica | | 分子名称: | PHOSPHATE ION, POTASSIUM ION, Peptidyl-prolyl cis-trans isomerase, ... | | 著者 | Bhatt, H, Pal, R.K, Tuteja, N, Bhavesh, N.S. | | 登録日 | 2012-05-02 | | 公開日 | 2013-05-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Structure of RNA-interacting Cyclophilin A-like protein from Piriformospora indica that provides salinity-stress tolerance in plants
Sci Rep, 3, 2013
|
|
3GSD
 
 | | 2.05 Angstrom structure of a divalent-cation tolerance protein (CutA) from Yersinia pestis | | 分子名称: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2009-03-26 | | 公開日 | 2009-04-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | 2.05 Angstrom Structure of a Divalent-cation Tolerance Protein (CutA) from Yersinia pestis
TO BE PUBLISHED
|
|
4URI
 
 | | Crystal structure of chitinase-like agglutinin RobpsCRA from Robinia pseudoacacia | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHITINASE-RELATED AGGLUTININ, CHLORIDE ION, ... | | 著者 | Sulzenbacher, G, Roig-Zamboni, V, Peumans, W.J, Henrissat, B, van Damme, E.J.M, Bourne, Y. | | 登録日 | 2014-06-30 | | 公開日 | 2015-03-11 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural Basis for Carbohydrate Binding Properties of a Plant Chitinase-Like Agglutinin with Conserved Catalytic Machinery.
J.Struct.Biol., 190, 2015
|
|
3HWG
 
 | |
4ZI6
 
 | |
4WM6
 
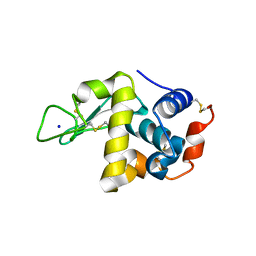 | |
3HZN
 
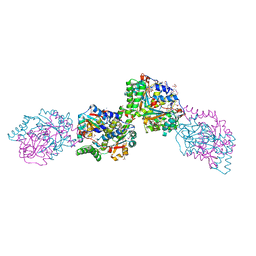 | | Structure of the Salmonella typhimurium nfnB dihydropteridine reductase | | 分子名称: | ACETATE ION, CHLORIDE ION, CITRATE ANION, ... | | 著者 | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Skarina, T, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2009-06-23 | | 公開日 | 2009-07-07 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure of the Salmonella typhimurium nfnB dihydropteridine reductase
TO BE PUBLISHED
|
|
3VD7
 
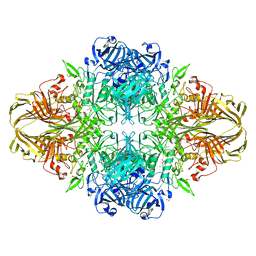 | | E. coli (lacZ) beta-galactosidase (N460S) in complex with galactotetrazole | | 分子名称: | (5R, 6S, 7S, ... | | 著者 | Wheatley, R.W, Kappelhoff, J.C, Hahn, J.N, Dugdale, M.L, Dutkoski, M.J, Tamman, S.D, Fraser, M.E, Huber, R.E. | | 登録日 | 2012-01-04 | | 公開日 | 2012-04-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.87 Å) | | 主引用文献 | Substitution for Asn460 cripples {beta}-galactosidase (Escherichia coli) by increasing substrate affinity and decreasing transition state stability.
Arch.Biochem.Biophys., 521, 2012
|
|
3I01
 
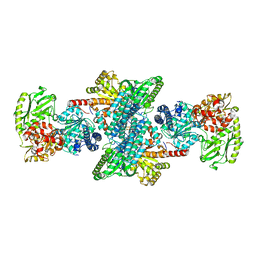 | | Native structure of bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase from Moorella thermoacetica, water-bound C-cluster. | | 分子名称: | ACETATE ION, COPPER (I) ION, Carbon monoxide dehydrogenase/acetyl-CoA synthase subunit alpha, ... | | 著者 | Kung, Y, Doukov, T.I, Drennan, C.L. | | 登録日 | 2009-06-24 | | 公開日 | 2009-09-01 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystallographic snapshots of cyanide- and water-bound C-clusters from bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase.
Biochemistry, 48, 2009
|
|
3I04
 
 | | Cyanide-bound structure of bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase from Moorella thermoacetica, cyanide-bound C-cluster | | 分子名称: | ACETATE ION, COPPER (I) ION, CYANIDE ION, ... | | 著者 | Kung, Y, Doukov, T.I, Drennan, C.L. | | 登録日 | 2009-06-24 | | 公開日 | 2009-09-01 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystallographic snapshots of cyanide- and water-bound C-clusters from bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase.
Biochemistry, 48, 2009
|
|
4WFX
 
 | |
5EPU
 
 | | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with cytosine at 1.06A. | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-AMINOPYRIMIDIN-2(1H)-ONE, ... | | 著者 | Prokofev, I.I, Lashkov, A.A, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | 登録日 | 2015-11-12 | | 公開日 | 2016-11-23 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.06 Å) | | 主引用文献 | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with cytosine at 1.06A.
To Be Published
|
|
