5LIT
 
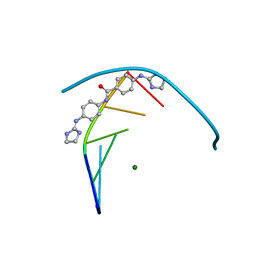 | | Structure of the DNA duplex d(AAATTT)2 with the potential antiparasitic drug 6XV at 1.25 A resolution | | 分子名称: | 4-((4,5-dihydro-1H-imidazol-2-yl)amino)-N-(4-((4,5-dihydro-1H-imidazol-2-yl)amino)phenyl)benzamide dihydrochloride, DNA (5'-D(*AP*AP*AP*TP*TP*T)-3'), MAGNESIUM ION | | 著者 | Millan, C.R, Dardonville, C, de Koning, H.P, Saperas, N, Lourdes Campos, J. | | 登録日 | 2016-07-15 | | 公開日 | 2017-06-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Functional and structural analysis of AT-specific minor groove binders that disrupt DNA-protein interactions and cause disintegration of the Trypanosoma brucei kinetoplast.
Nucleic Acids Res., 45, 2017
|
|
5LP3
 
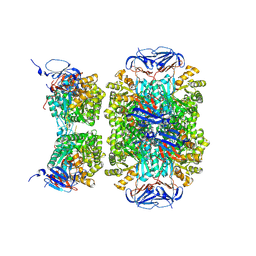 | |
5LJ3
 
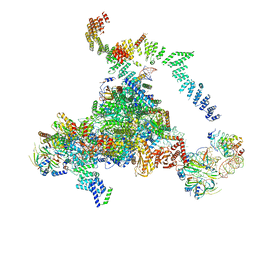 | | Structure of the core of the yeast spliceosome immediately after branching | | 分子名称: | CEF1, CLF1, CWC15, ... | | 著者 | Galej, W.P, Wilkinson, M.F, Fica, S.M, Oubridge, C, Newman, A.J, Nagai, K. | | 登録日 | 2016-07-17 | | 公開日 | 2016-08-03 | | 最終更新日 | 2019-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Cryo-EM structure of the spliceosome immediately after branching.
Nature, 537, 2016
|
|
3W31
 
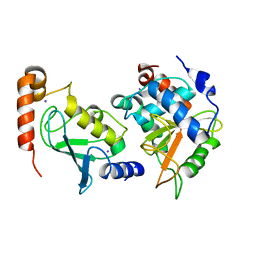 | | Structual basis for the recognition of Ubc13 by the Shigella flexneri effector OspI | | 分子名称: | IODIDE ION, ORF169b, Ubiquitin-conjugating enzyme E2 N | | 著者 | Nishide, A, Kim, M, Takagi, K, Sasakawa, C, Mizushima, T. | | 登録日 | 2012-12-07 | | 公開日 | 2013-03-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.96 Å) | | 主引用文献 | Structural basis for the recognition of Ubc13 by the Shigella flexneri effector OspI.
J.Mol.Biol., 425, 2013
|
|
8JOJ
 
 | | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 in complex with 9,11-epoxy-17-hydroxypregn-4-ene-3,20-dione actate | | 分子名称: | 3-ketosteroid dehydrogenase, 9,11-epoxy-17-hydroxypregn-4-ene-3,20-dione actate, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Hu, Y.L, Li, X, Cheng, X.Y, Song, S.K, Su, Z.D. | | 登録日 | 2023-06-07 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.819 Å) | | 主引用文献 | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 in complex with 9,11-epoxy-17-hydroxypregn-4-ene-3,20-dione actate
To Be Published
|
|
4HXN
 
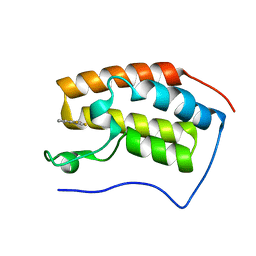 | | Brd4 Bromodomain 1 complex with 4-(2-FLUOROPHENYL)-1,3-THIAZOL-2(3H)-ONE inhibitor | | 分子名称: | 4-(2-fluorophenyl)-1,3-thiazol-2(3H)-one, Bromodomain-containing protein 4 | | 著者 | Chen, T.T, Cao, D.Y, Chen, W.Y, Xiong, B, Shen, J.K, Xu, Y.C. | | 登録日 | 2012-11-12 | | 公開日 | 2013-04-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Fragment-Based Drug Discovery of 2-Thiazolidinones as Inhibitors of the Histone Reader BRD4 Bromodomain.
J.Med.Chem., 56, 2013
|
|
4Q7A
 
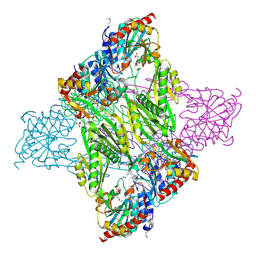 | | Crystal Structure of N-acetyl-ornithine/N-acetyl-lysine Deacetylase from Sphaerobacter thermophilus | | 分子名称: | CHLORIDE ION, GLYCEROL, N-acetyl-ornithine/N-acetyl-lysine deacetylase, ... | | 著者 | Kim, Y, Tesar, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-04-24 | | 公開日 | 2014-07-02 | | 実験手法 | X-RAY DIFFRACTION (2.048 Å) | | 主引用文献 | Crystal Structure of N-acetyl-ornithine/N-acetyl-lysine Deacetylase from
Sphaerobacter thermophilus
To be Published
|
|
6B0D
 
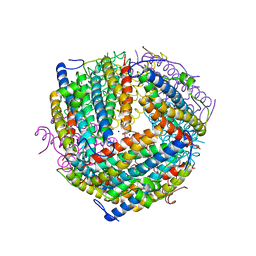 | | An E. coli DPS protein from ferritin superfamily | | 分子名称: | DNA protection during starvation protein, FORMIC ACID, SODIUM ION | | 著者 | Rui, W, Ruslan, S, Ronan, K, Adam, J.S. | | 登録日 | 2017-09-14 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | SIMBAD: a sequence-independent molecular-replacement pipeline.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
8D7F
 
 | |
6B0Z
 
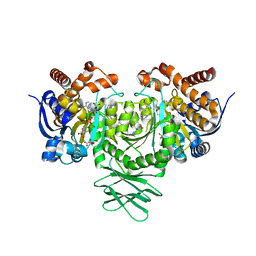 | | IDH1 R132H mutant in complex with IDH305 | | 分子名称: | (4R)-4-[(1S)-1-fluoroethyl]-3-[2-({(1S)-1-[4-methyl-2'-(trifluoromethyl)[3,4'-bipyridin]-6-yl]ethyl}amino)pyrimidin-4-yl]-1,3-oxazolidin-2-one, CITRATE ANION, Isocitrate dehydrogenase [NADP] cytoplasmic, ... | | 著者 | Xie, X, Kulathila, R. | | 登録日 | 2017-09-15 | | 公開日 | 2017-11-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.334 Å) | | 主引用文献 | Discovery and Evaluation of Clinical Candidate IDH305, a Brain Penetrant Mutant IDH1 Inhibitor.
ACS Med Chem Lett, 8, 2017
|
|
3NXR
 
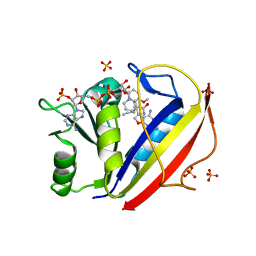 | | Perferential Selection of Isomer Binding from Chiral Mixtures: Alternate Binding Modes Observed for the E- and Z-isomers of a Series of 5-Substituted 2,4-Diaminofuro[2,3-d]pyrimidines as Ternary Complexes with NADPH and Human Dihydrofolate Reductase | | 分子名称: | 5-[(1E)-2-(2-methoxyphenyl)-4-methylpent-1-en-1-yl]furo[2,3-d]pyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Cody, V. | | 登録日 | 2010-07-14 | | 公開日 | 2010-12-01 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Preferential selection of isomer binding from chiral mixtures: alternate binding modes observed for the E and Z isomers of a series of 5-substituted 2,4-diaminofuro[2,3-d]pyrimidines as ternary complexes with NADPH and human dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3NXX
 
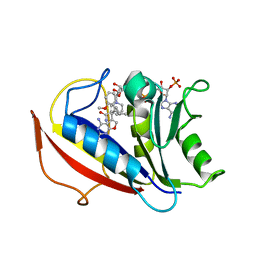 | | Preferential Selection of Isomer Binding from Chiral Mixtures: Alternate Binding Modes Observed for the E- and Z-isomers of a Series of 5-Substituted 2,4-Diaminofuro-2,3-d]pyrimidines as Ternary Complexes with NADPH and Human Dihydrofolate Reductase | | 分子名称: | 5-[(1E)-2-(2-methoxyphenyl)-4-methylpent-1-en-1-yl]furo[2,3-d]pyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Cody, V. | | 登録日 | 2010-07-14 | | 公開日 | 2010-12-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Preferential selection of isomer binding from chiral mixtures: alternate binding modes observed for the E and Z isomers of a series of 5-substituted 2,4-diaminofuro[2,3-d]pyrimidines as ternary complexes with NADPH and human dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
8A4O
 
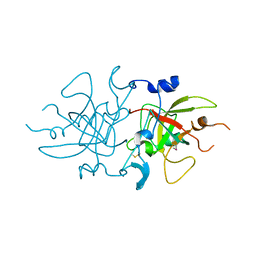 | |
1S40
 
 | | SOLUTION STRUCTURE OF THE CDC13 DNA-BINDING DOMAIN COMPLEXED WITH A SINGLE-STRANDED TELOMERIC DNA 11-MER | | 分子名称: | 5'-D(*GP*TP*GP*TP*GP*GP*GP*TP*GP*TP*G)-3', Cell division control protein 13 | | 著者 | Mitton-Fry, R.M, Anderson, E.M, Theobald, D.L, Glustrom, L.W, Wuttke, D.S. | | 登録日 | 2004-01-14 | | 公開日 | 2004-05-04 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for telomeric single-stranded DNA recognition by yeast Cdc13
J.Mol.Biol., 338, 2004
|
|
1ILQ
 
 | |
8J51
 
 | |
4QGS
 
 | |
8A14
 
 | |
8J50
 
 | |
1BHJ
 
 | |
8J52
 
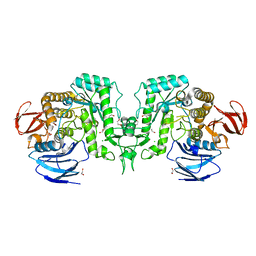 | | Crystal structure of Flavihumibacter petaseus GH31 alpha-galactosidase mutant D304A in complex with alpha-1,4-galactobiose | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, GH31 alpha-galactosidase, ... | | 著者 | Ikegaya, M, Miyazaki, T. | | 登録日 | 2023-04-21 | | 公開日 | 2023-07-26 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure-function analysis of bacterial GH31 alpha-galactosidases specific for alpha-(1→4)-galactobiose.
Febs J., 290, 2023
|
|
8J53
 
 | |
3WAL
 
 | | Crystal structure of human LC3A_2-121 | | 分子名称: | D-MALATE, Microtubule-associated proteins 1A/1B light chain 3A | | 著者 | Suzuki, H, Tabata, K, Morita, E, Kawasaki, M, Kato, R, Dobson, R.C.J, Yoshimori, T, Wakatsuki, S. | | 登録日 | 2013-05-06 | | 公開日 | 2013-12-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism.
Structure, 22, 2014
|
|
1ATI
 
 | | CRYSTAL STRUCTURE OF GLYCYL-TRNA SYNTHETASE FROM THERMUS THERMOPHILUS | | 分子名称: | GLYCYL-TRNA SYNTHETASE, GLYCYL-tRNA SYNTHETASE | | 著者 | Logan, D.T, Mazauric, M.-H, Kern, D, Moras, D. | | 登録日 | 1996-04-23 | | 公開日 | 1997-07-07 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Crystal structure of glycyl-tRNA synthetase from Thermus thermophilus.
EMBO J., 14, 1995
|
|
3O3P
 
 | | Crystal structure of R. xylanophilus MpgS in complex with GDP-Mannose | | 分子名称: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, ... | | 著者 | Macedo-Ribeiro, S, Pereira, P.J.B, Empadinhas, N, da Costa, M.S. | | 登録日 | 2010-07-25 | | 公開日 | 2010-11-03 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.529 Å) | | 主引用文献 | Functional and structural characterization of a novel mannosyl-3-phosphoglycerate synthase from Rubrobacter xylanophilus reveals its dual substrate specificity
Mol.Microbiol., 79, 2011
|
|
