7P5B
 
 | | Variant Surface Glycoprotein 3 (VSG3, MiTat1.3, VSG224) mutant (serine 319 to alanine), single O-linked glycosylated at Ser317 | | 分子名称: | Variant surface glycoprotein, alpha-D-glucopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Gkeka, A, Aresta-Branco, F, Stebbins, C.E, Papavasiliou, F.N. | | 登録日 | 2021-07-14 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.13 Å) | | 主引用文献 | Immunodominant surface epitopes power immune evasion in the African trypanosome.
Cell Rep, 42, 2023
|
|
8XYZ
 
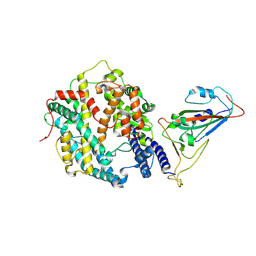 | | The structure of fox ACE2 and PT RBD complex | | 分子名称: | Angiotensin-converting enzyme, Signal peptide, Spike protein S1, ... | | 著者 | sun, J.Q. | | 登録日 | 2024-01-20 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.96 Å) | | 主引用文献 | The binding and structural basis of fox ACE2 to RBDs from different sarbecoviruses.
Virol Sin, 2024
|
|
9F3D
 
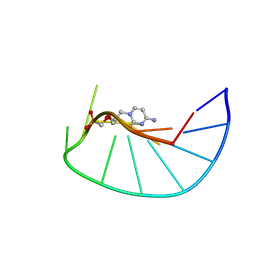 | |
8VMQ
 
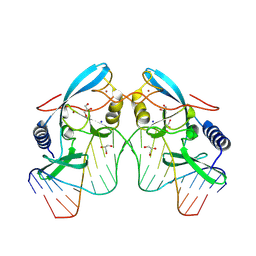 | |
8VMO
 
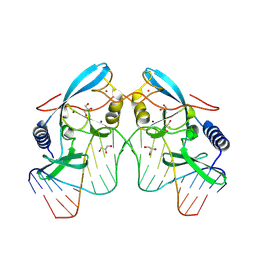 | |
8VMY
 
 | |
8VN0
 
 | |
9EXW
 
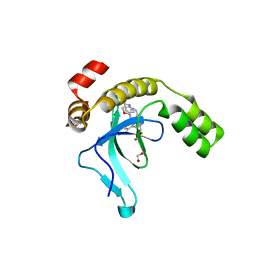 | | Crystal structure of the PWWP1 domain of NSD2 bound by compound 17. | | 分子名称: | 1,2-ETHANEDIOL, 7-[3-methyl-5-[2-methyl-5-[(pyridin-3-ylamino)methyl]phenyl]imidazol-4-yl]-4~{H}-1,4-benzoxazin-3-one, Histone-lysine N-methyltransferase NSD2 | | 著者 | Collie, G.W. | | 登録日 | 2024-04-09 | | 公開日 | 2024-05-29 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | Identification of Novel Potent NSD2-PWWP1 Ligands Using Structure-Based Design and Computational Approaches.
J.Med.Chem., 67, 2024
|
|
9B5P
 
 | |
9B5W
 
 | |
8X22
 
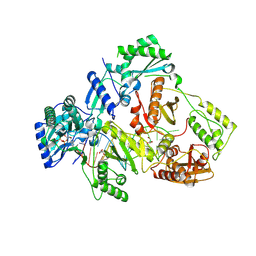 | | HIV-1 reverse transcriptase mutant Q151M/Y115F/F116Y/L74V:DNA:dGTP ternary complex | | 分子名称: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | 著者 | Yasutake, Y, Hattori, S.I, Mitsuya, H. | | 登録日 | 2023-11-09 | | 公開日 | 2024-07-17 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Deviated binding of anti-HBV nucleoside analog E-CFCP-TP to the reverse transcriptase active site attenuates the effect of drug-resistant mutations.
Sci Rep, 14, 2024
|
|
7N4D
 
 | |
9B5T
 
 | |
7N6S
 
 | |
7N7S
 
 | |
7NOS
 
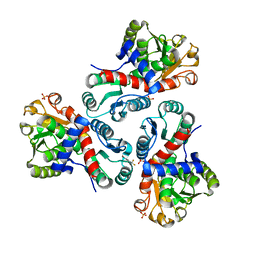 | | Crystal structure of Mycobacterium tuberculosis ArgF in complex with 4-bromo-6-(trifluoromethyl)-1H-benzo[d]imidazole. | | 分子名称: | 4-bromanyl-6-(trifluoromethyl)-1~{H}-benzimidazole, Ornithine carbamoyltransferase, PHOSPHATE ION | | 著者 | Mendes, V, Gupta, P, Burgess, A, Sebastian-Perez, V, Cattermole, E, Meghir, C, Blundell, T.L. | | 登録日 | 2021-02-25 | | 公開日 | 2021-06-30 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
4YM8
 
 | |
7NNF
 
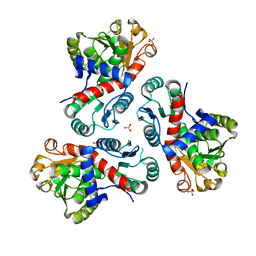 | | Crystal structure of Mycobacterium tuberculosis ArgF in apo form. | | 分子名称: | Ornithine carbamoyltransferase, PHOSPHATE ION | | 著者 | Mendes, V, Gupta, P, Burgess, A, Sebastian-Perez, V, Cattermole, E, Meghir, C, Blundell, T.L. | | 登録日 | 2021-02-24 | | 公開日 | 2021-06-30 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7OL2
 
 | |
7NOU
 
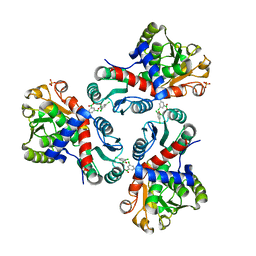 | | Crystal structure of Mycobacterium tuberculosis ArgF in complex with (3,5-dichlorophenyl)boronic acid. | | 分子名称: | Ornithine carbamoyltransferase, PHOSPHATE ION, [3,5-bis(chloranyl)phenyl]-oxidanyl-oxidanylidene-boron | | 著者 | Mendes, V, Gupta, P, Burgess, A, Sebastian-Perez, V, Cattermole, E, Meghir, C, Blundell, T.L. | | 登録日 | 2021-02-25 | | 公開日 | 2021-06-30 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
6X2B
 
 | |
7NNY
 
 | | Crystal structure of Mycobacterium tuberculosis ArgF in complex with naphthalen-1-ol. | | 分子名称: | 1-NAPHTHOL, Ornithine carbamoyltransferase, PHOSPHATE ION | | 著者 | Mendes, V, Gupta, P, Burgess, A, Sebastian-Perez, V, Cattermole, E, Meghir, C, Blundell, T.L. | | 登録日 | 2021-02-25 | | 公開日 | 2021-06-30 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NNZ
 
 | | Crystal structure of Mycobacterium tuberculosis ArgF in complex with 5-methyl-4-phenylthiazol-2-amine. | | 分子名称: | 5-methyl-4-phenyl-1,3-thiazol-2-amine, Ornithine carbamoyltransferase, PHOSPHATE ION | | 著者 | Mendes, V, Gupta, P, Burgess, A, Sebastian-Perez, V, Cattermole, E, Meghir, C, Blundell, T.L. | | 登録日 | 2021-02-25 | | 公開日 | 2021-06-30 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NP0
 
 | | Crystal structure of Mycobacterium tuberculosis ArgF in complex with (4-nitrophenyl)boronic acid. | | 分子名称: | Ornithine carbamoyltransferase, PHOSPHATE ION, p-nitrophenylboronic acid | | 著者 | Mendes, V, Gupta, P, Burgess, A, Sebastian-Perez, V, Cattermole, E, Meghir, C, Blundell, T.L. | | 登録日 | 2021-02-26 | | 公開日 | 2021-06-30 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
6WPB
 
 | |
