4RNY
 
 | | Structure of Helicobacter pylori Csd3 from the orthorhombic crystal | | 分子名称: | Conserved hypothetical secreted protein, GLYCEROL, SULFATE ION, ... | | 著者 | An, D.R, Kim, H.S, Kim, J, Im, H.N, Yoon, H.J, Yoon, J.Y, Jang, J.Y, Hesek, D, Lee, M, Mobashery, S, Kim, S.-J, Lee, B.I, Suh, S.W. | | 登録日 | 2014-10-27 | | 公開日 | 2015-03-11 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of Csd3 from Helicobacter pylori, a cell shape-determining metallopeptidase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4RT0
 
 | | Structure of the Alg44 PilZ domain from Pseudomonas aeruginosa PAO1 in complex with c-di-GMP | | 分子名称: | 1,2-ETHANEDIOL, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Alginate biosynthesis protein Alg44 | | 著者 | Whitfield, G.B, Whitney, J.C. | | 登録日 | 2014-11-12 | | 公開日 | 2015-04-08 | | 最終更新日 | 2015-06-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Dimeric c-di-GMP Is Required for Post-translational Regulation of Alginate Production in Pseudomonas aeruginosa.
J.Biol.Chem., 290, 2015
|
|
4TU0
 
 | | CRYSTAL STRUCTURE OF CHIKUNGUNYA VIRUS NSP3 MACRO DOMAIN IN COMPLEX WITH A 2'-5' OLIGOADENYLATE TRIMER | | 分子名称: | 2'-5' OLIGOADENYLATE TRIMER, DI(HYDROXYETHYL)ETHER, Non-structural polyprotein 3 | | 著者 | Morin, B, Ferron, f.p, Malet, h, Coutard, b, Canard, b. | | 登録日 | 2014-06-23 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF CHIKUNGUNYA VIRUS NSP3 MACRO DOMAIN IN COMPLEX WITH A 2'-5' OLIGOADENYLATE TRIMER
TO BE PUBLISHED
|
|
5TXM
 
 | | STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING DDATP | | 分子名称: | 1,2-ETHANEDIOL, 2',3'-dideoxyadenosine triphosphate, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*G)-3'), ... | | 著者 | Das, K, Martinez, S.M, Arnold, E. | | 登録日 | 2016-11-17 | | 公開日 | 2017-04-05 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural Insights into HIV Reverse Transcriptase Mutations Q151M and Q151M Complex That Confer Multinucleoside Drug Resistance.
Antimicrob. Agents Chemother., 61, 2017
|
|
2P3D
 
 | | Crystal Structure of the multi-drug resistant mutant subtype F HIV protease complexed with TL-3 inhibitor | | 分子名称: | Pol protein, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate | | 著者 | Sanches, M, Krauchenco, S, Martins, N.H, Gustchina, A, Wlodawer, A, Polikarpov, I. | | 登録日 | 2007-03-08 | | 公開日 | 2007-04-24 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural Characterization of B and non-B Subtypes of HIV-Protease: Insights into the Natural Susceptibility to Drug Resistance Development.
J.Mol.Biol., 369, 2007
|
|
5U38
 
 | | Crystal structure of native lectin from Platypodium elegans seeds (PELa) complexed with Man1-3Man-OMe. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Lectin, ... | | 著者 | Silva, I.B, Araripe, D.A, Neco, A.H.B, Pinto-Junior, V.R, Osterne, V.J.S, Santiago, M.Q, Silva-Filho, J.C, Leal, R.B, Rocha, C.R.C, Nascimento, K.S, Cavada, B.S. | | 登録日 | 2016-12-01 | | 公開日 | 2017-10-04 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural studies and nociceptive activity of a native lectin from Platypodium elegans seeds (nPELa).
Int. J. Biol. Macromol., 107, 2018
|
|
2XH4
 
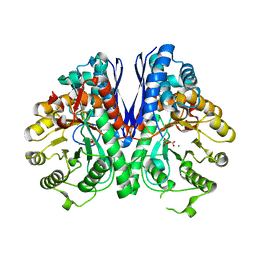 | |
4MMT
 
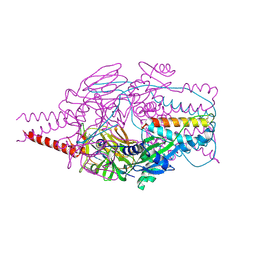 | | Crystal Structure of Prefusion-stabilized RSV F Variant DS-Cav1 at pH 9.5 | | 分子名称: | Fusion glycoprotein F1 fused with Fibritin trimerization domain, Fusion glycoprotein F2 | | 著者 | Joyce, M.G, Mclellan, J.S, Stewart-Jones, G.B.E, Sastry, M, Yang, Y, Graham, B.S, Kwong, P.D. | | 登録日 | 2013-09-09 | | 公開日 | 2013-11-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus.
Science, 342, 2013
|
|
1L19
 
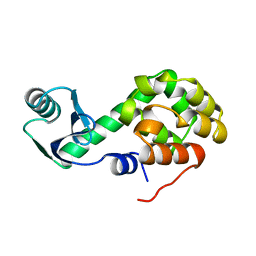 | |
1L25
 
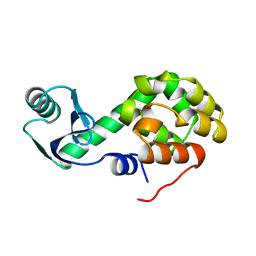 | |
1L22
 
 | |
1L31
 
 | |
4U78
 
 | | Octameric RNA duplex soaked in copper(II)chloride | | 分子名称: | CALCIUM ION, COPPER (II) ION, RNA (5'-R(*UP*CP*GP*UP*AP*CP*GP*A)-3') | | 著者 | Schaffer, M.F, Spingler, B, Schnabl, J, Peng, G, Olieric, V, Sigel, R.K.O. | | 登録日 | 2014-07-30 | | 公開日 | 2015-08-26 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.501 Å) | | 主引用文献 | The X-ray Structures of Six Octameric RNA Duplexes in the Presence of Different Di- and Trivalent Cations.
Int J Mol Sci, 17, 2016
|
|
5V5I
 
 | |
5UZU
 
 | | Immune evasion by a Staphylococcal Peroxidase Inhibitor that blocks myeloperoxidase | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | de Jong, N, Geisbrecht, B.V, van Strijp, J, Haas, P, Nijland, R, Ramyar, K, Fevre, C, Guerra, F, Voyich-Kane, J, van Kessel, C, Garcia, B. | | 登録日 | 2017-02-27 | | 公開日 | 2017-07-26 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.403 Å) | | 主引用文献 | Immune evasion by a staphylococcal inhibitor of myeloperoxidase.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1L23
 
 | |
5V8V
 
 | | Crystal Structure of Human Renin in Complex with a biphenylpipderidinylcarbinol | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin, methyl [(4S)-4-(3'-ethyl-6-fluoro[1,1'-biphenyl]-2-yl)-4-hydroxy-4-{(3R)-1-[4-(methylamino)butanoyl]piperidin-3-yl}butyl]carbamate | | 著者 | Concha, N, Zhao, B. | | 登録日 | 2017-03-22 | | 公開日 | 2017-10-18 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Discovery of renin inhibitors containing a simple aspartate binding moiety that imparts reduced P450 inhibition.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
1QBB
 
 | | BACTERIAL CHITOBIASE COMPLEXED WITH CHITOBIOSE (DINAG) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITOBIASE, SULFATE ION | | 著者 | Tews, I, Perrakis, A, Oppenheim, A, Dauter, Z, Wilson, K.S, Vorgias, C.E. | | 登録日 | 1996-06-07 | | 公開日 | 1997-02-12 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease.
Nat.Struct.Biol., 3, 1996
|
|
4B4F
 
 | | Thermobifida fusca Cel6B(E3) co-crystallized with cellobiose | | 分子名称: | BETA-1,4-EXOCELLULASE, CALCIUM ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | 著者 | Sandgren, M, Wu, M, Stahlberg, J, Karkehabadi, S, Mitchinson, C, Kelemen, B.R, Larenas, E.A, Hansson, H. | | 登録日 | 2012-07-30 | | 公開日 | 2012-12-19 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The Structure of a Bacterial Cellobiohydrolase: The Catalytic Core of the Thermobifida Fusca Family Gh6 Cellobiohydrolase Cel6B.
J.Mol.Biol., 425, 2013
|
|
4AZG
 
 | | Differential inhibition of the tandem GH20 catalytic modules in the pneumococcal exo-beta-D-N-acetylglucosaminidase, StrH | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, BETA-N-ACETYLHEXOSAMINIDASE, ... | | 著者 | Pluvinage, B, Stubbs, K.A, Vocadlo, D.J, Boraston, A.B. | | 登録日 | 2012-06-25 | | 公開日 | 2013-07-10 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Inhibition of the Family 20 Glycoside Hydrolase Catalytic Modules in the Streptococcus Pneumoniae Exo-Beta-D-N-Acetylglucosaminidase, Strh.
Org.Biomol.Chem., 11, 2013
|
|
2QEF
 
 | | X-ray structure of 7-deaza-dG and Z3dU modified duplex CGCGAATXCZCG | | 分子名称: | DNA (5'-D(*DCP*DGP*DCP*DGP*DAP*DAP*DTP*(ZDU)P*DCP*(7GU)P*DCP*DG)-3') | | 著者 | Wang, F, Li, F, Ganguly, M, Marky, L.A, Gold, B, Egli, M, Stone, M.P. | | 登録日 | 2007-06-25 | | 公開日 | 2008-05-06 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | A bridging water anchors the tethered 5-(3-aminopropyl)-2'-deoxyuridine amine in the DNA major groove proximate to the N+2 C.G base pair: implications for formation of interstrand 5'-GNC-3' cross-links by nitrogen mustards.
Biochemistry, 47, 2008
|
|
4AZI
 
 | | Differential inhibition of the tandem GH20 catalytic modules in the pneumococcal exo-beta-D-N-acetylglucosaminidase, StrH | | 分子名称: | 1,2-ETHANEDIOL, BETA-N-ACETYLHEXOSAMINIDASE, BICINE, ... | | 著者 | Pluvinage, B, Stubbs, K.A, Vocadlo, D.J, Boraston, A.B. | | 登録日 | 2012-06-26 | | 公開日 | 2013-07-10 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Inhibition of the Family 20 Glycoside Hydrolase Catalytic Modules in the Streptococcus Pneumoniae Exo-Beta-D-N-Acetylglucosaminidase, Strh.
Org.Biomol.Chem., 11, 2013
|
|
3FAR
 
 | |
3PFD
 
 | |
2XH7
 
 | |
