6G6Q
 
 | | Crystal structure of the computationally designed Ika4 protein | | 分子名称: | Ika4 | | 著者 | Noguchi, H, Addy, C, Simoncini, D, Van Meervelt, L, Schiex, T, Zhang, K.Y.J, Tame, J.R.H, Voet, A.R.D. | | 登録日 | 2018-04-01 | | 公開日 | 2018-11-28 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Computational design of symmetrical eight-bladed beta-propeller proteins.
IUCrJ, 6, 2019
|
|
7DMR
 
 | | Crystal structure of potassium induced heme modification in yak lactoperoxidase at 2.20 A resolution | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Singh, P.K, Rani, C, Sharma, P, Sharma, S, Singh, T.P. | | 登録日 | 2020-12-06 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Potassium-induced partial inhibition of lactoperoxidase: structure of the complex of lactoperoxidase with potassium ion at 2.20 angstrom resolution.
J.Biol.Inorg.Chem., 26, 2021
|
|
6VH1
 
 | | 2.30 A resolution structure of MERS 3CL protease in complex with inhibitor 6h | | 分子名称: | N~2~-{[(4,4-difluorocyclohexyl)oxy]carbonyl}-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide, Orf1a protein | | 著者 | Lovell, S, Battaile, K.P, Kashipathy, M.M, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | 登録日 | 2020-01-09 | | 公開日 | 2020-08-12 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
7DS7
 
 | | The Crystal Structure of Leaf-branch compost cutinase from Biortus. | | 分子名称: | CITRIC ACID, GLYCEROL, IMIDAZOLE, ... | | 著者 | Wang, F, Lv, Z, Cheng, W, Lin, D, Chu, F, Xu, X, Tan, J. | | 登録日 | 2020-12-30 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | The Crystal Structure of Leaf-branch compost cutinase from Biortus.
To Be Published
|
|
6G76
 
 | |
5KKH
 
 | | 2.1-Angstrom In situ Mylar structure of bacteriorhodopsin from Haloquadratum walsbyi (HwBR) at 100 K | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin-I, ... | | 著者 | Broecker, J, Ernst, O.P. | | 登録日 | 2016-06-21 | | 公開日 | 2017-02-15 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.125 Å) | | 主引用文献 | A Versatile System for High-Throughput In Situ X-ray Screening and Data Collection of Soluble and Membrane-Protein Crystals.
Cryst Growth Des, 16, 2016
|
|
5HTS
 
 | | Crystal structure of shaft pilin spaA from Lactobacillus rhamnosus GG - D295N mutant | | 分子名称: | Cell surface protein SpaA | | 著者 | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | 登録日 | 2016-01-27 | | 公開日 | 2016-07-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
6FTW
 
 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-048 | | 分子名称: | 1,2-ETHANEDIOL, 3-{5-[(4aR,8aS)-3-cycloheptyl-4-oxo-3,4,4a,5,8,8a-hexahydrophthalazin-1-yl]-2-methoxyphenyl}prop-2-ynamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Singh, A.K, Brown, D.G. | | 登録日 | 2018-02-24 | | 公開日 | 2019-03-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Alkynamide phthalazinones as a new class of TbrPDEB1 inhibitors.
Bioorg.Med.Chem., 27, 2019
|
|
5TZB
 
 | | Burkholderia sp. beta-aminopeptidase | | 分子名称: | CALCIUM ION, D-aminopeptidase | | 著者 | McGowan, S, Drinkwater, N, John, M, Dumsday, G. | | 登録日 | 2016-11-21 | | 公開日 | 2017-07-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.977 Å) | | 主引用文献 | Crystal structure of a beta-aminopeptidase from an Australian Burkholderia sp.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
6MPS
 
 | | TagT bound to LIIa-WTA | | 分子名称: | 2-(acetylamino)-4-O-[2-(acetylamino)-2-deoxy-beta-D-mannopyranosyl]-2-deoxy-1-O-[(S)-{[(R)-{[(2Z,6Z,10Z,14E,18E)-3,7,11,15,19,23-hexamethyltetracosa-2,6,10,14,18,22-hexaen-1-yl]oxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]-alpha-D-glucopyranose, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Owens, T.W, Schaefer, K, Kahne, D, Walker, S. | | 登録日 | 2018-10-08 | | 公開日 | 2018-10-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Substrate Preferences Establish the Order of Cell Wall Assembly in Staphylococcus aureus.
J. Am. Chem. Soc., 140, 2018
|
|
5U0C
 
 | |
6VHU
 
 | | Klebsiella oxytoca NpsA N-terminal subdomain in space group P21 | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BROMIDE ION, CHLORIDE ION, ... | | 著者 | Kreitler, D.F, Gulick, A.M. | | 登録日 | 2020-01-10 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Biosynthesis, Mechanism of Action, and Inhibition of the Enterotoxin Tilimycin Produced by the Opportunistic PathogenKlebsiella oxytoca.
Acs Infect Dis., 6, 2020
|
|
7CY3
 
 | |
6XPU
 
 | |
5DN5
 
 | | Structure of a C-terminally truncated glycoside hydrolase domain from Salmonella typhimurium FlgJ | | 分子名称: | CHLORIDE ION, IODIDE ION, Peptidoglycan hydrolase FlgJ, ... | | 著者 | Zaloba, P, Bailey-Elkin, B.A, Mark, B.L. | | 登録日 | 2015-09-09 | | 公開日 | 2016-02-24 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structural and Biochemical Insights into the Peptidoglycan Hydrolase Domain of FlgJ from Salmonella typhimurium.
Plos One, 11, 2016
|
|
7CY9
 
 | |
5L7I
 
 | | Structure of human Smoothened in complex with Vismodegib | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-chloranyl-~{N}-(4-chloranyl-3-pyridin-2-yl-phenyl)-4-methylsulfonyl-benzamide, SODIUM ION, ... | | 著者 | Byrne, E.X.B, Sircar, R, Miller, P.S, Hedger, G, Luchetti, G, Nachtergaele, S, Tully, M.D, Mydock-McGrane, L, Covey, D.F, Rambo, R.F, Sansom, M.S.P, Newstead, S, Rohatgi, R, Siebold, C. | | 登録日 | 2016-06-03 | | 公開日 | 2016-07-20 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structural basis of Smoothened regulation by its extracellular domains.
Nature, 535, 2016
|
|
5HVI
 
 | |
6XQP
 
 | | Structure of human D462-E4 TCR in complex with human MR1-5-OP-RU | | 分子名称: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, BROMIDE ION, Beta-2-microglobulin, ... | | 著者 | Awad, W, Rossjohn, J. | | 登録日 | 2020-07-10 | | 公開日 | 2020-08-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Atypical TRAV1-2 - T cell receptor recognition of the antigen-presenting molecule MR1.
J.Biol.Chem., 295, 2020
|
|
5KMB
 
 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) H112N mutant nucleoside L-Trp phosphoramidate substrate complex | | 分子名称: | CHLORIDE ION, Histidine triad nucleotide-binding protein 1, [(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-~{N}-[(2~ {S})-3-(1~{H}-indol-3-yl)-1-(methylamino)-1-oxidanylidene-propan-2-yl]phosphonamidic acid | | 著者 | Maize, K.M, Finzel, B.C. | | 登録日 | 2016-06-26 | | 公開日 | 2017-06-28 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | A Crystal Structure Based Guide to the Design of Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) Activated ProTides.
Mol. Pharm., 14, 2017
|
|
8OMC
 
 | |
5KMP
 
 | |
5L8G
 
 | | Crystal structure of Rhodospirillum rubrum Rru_A0973 mutant H65A | | 分子名称: | CALCIUM ION, Uncharacterized protein | | 著者 | He, D, Hughes, S, Vanden-Hehir, S, Georgiev, A, Altenbach, K, Tarrant, E, Mackay, C.L, Waldron, K.J, Clarke, D.J, Marles-Wright, J. | | 登録日 | 2016-06-07 | | 公開日 | 2016-08-31 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.974 Å) | | 主引用文献 | Structural characterization of encapsulated ferritin provides insight into iron storage in bacterial nanocompartments.
Elife, 5, 2016
|
|
8KI1
 
 | | Crystal structure of the holo form of the hemophore HasA from Pseudomonas protegens Pf-5 | | 分子名称: | GLYCEROL, Heme acquisition protein HasAp, PHOSPHATE ION, ... | | 著者 | Shisaka, Y, Inaba, H, Sugimoto, H, Shoji, O. | | 登録日 | 2023-08-22 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Heme-substituted protein assembly bridged by synthetic porphyrin: achieving controlled configuration while maintaining rotational freedom.
Rsc Adv, 14, 2024
|
|
6XQQ
 
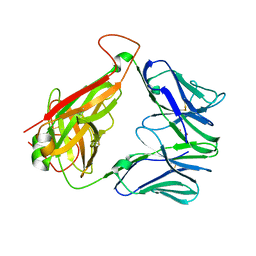 | | Structure of human D462-E4 TCR | | 分子名称: | GLYCEROL, TRAV12-2 alpha chain, TRBV29-1 | | 著者 | Awad, W, Rossjohn, J. | | 登録日 | 2020-07-10 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Atypical TRAV1-2 - T cell receptor recognition of the antigen-presenting molecule MR1.
J.Biol.Chem., 295, 2020
|
|
