2RJ2
 
 | | Crystal Structure of the Sugar Recognizing SCF Ubiquitin Ligase at 1.7 Resolution | | 分子名称: | CHLORIDE ION, F-box only protein 2, NICKEL (II) ION | | 著者 | Vaijayanthimala, S, Velmurugan, D, Mizushima, T, Yamane, T, Yoshida, Y, Tanaka, K. | | 登録日 | 2007-10-14 | | 公開日 | 2008-10-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal Structure of the Sugar Recognizing SCF Ubiquitin Ligase at 1.7 Resolution
To be Published
|
|
2RMW
 
 | |
5J4F
 
 | | Crystal structure of the N-terminally His6-tagged HP0902, an uncharacterized protein from Helicobacter pylori 26695 | | 分子名称: | Uncharacterized protein | | 著者 | Sim, D.-W, Lee, W.-C, Kim, H.Y, Kim, J.-H, Won, H.-S. | | 登録日 | 2016-04-01 | | 公開日 | 2017-02-08 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural identification of the lipopolysaccharide-binding capability of a cupin-family protein from Helicobacter pylori
FEBS Lett., 590, 2016
|
|
2RNX
 
 | | The Structural Basis for Site-Specific Lysine-Acetylated Histone Recognition by the Bromodomains of the HUman Transcriptional Co-Activators PCAF and CBP | | 分子名称: | Histone H3, Histone acetyltransferase PCAF | | 著者 | Zeng, L, Zhang, Q, Gerona-Navarro, G, Zhou, M.M. | | 登録日 | 2008-02-03 | | 公開日 | 2008-05-06 | | 最終更新日 | 2023-11-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Basis of Site-Specific Histone Recognition by the Bromodomains of Human Coactivators PCAF and CBP/p300
Structure, 16, 2008
|
|
5JAB
 
 | |
5J61
 
 | |
7BYW
 
 | |
2RLL
 
 | | CCR5 Nt(7-15) | | 分子名称: | 9-mer from C-C chemokine receptor type 5 | | 著者 | Bewley, C.A, Lam, S.N. | | 登録日 | 2007-07-21 | | 公開日 | 2007-09-25 | | 最終更新日 | 2023-11-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4
Science, 317, 2007
|
|
2RVO
 
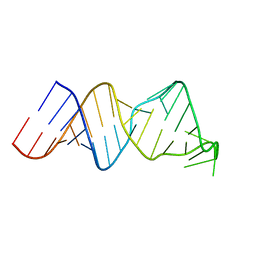 | | Solution structure of a reverse transcriptase recognition site of a LINE RNA from zebrafish | | 分子名称: | RNA (34-MER) | | 著者 | Otsu, M, Norose, N, Arai, N, Terao, R, Kajikawa, M, Okada, N, Kawai, G. | | 登録日 | 2016-02-03 | | 公開日 | 2017-02-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a reverse transcriptase recognition site of a LINE RNA from zebrafish.
J. Biochem., 162, 2017
|
|
6MIQ
 
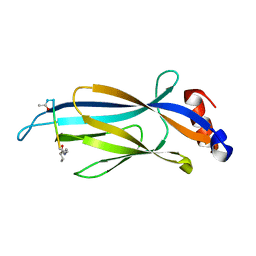 | | Crystal structure of Taf14 YEATS domain in complex with histone H3K9bu | | 分子名称: | Histone H3K9bu, Transcription initiation factor TFIID subunit 14 | | 著者 | Klein, B.J, Andrews, F.H, Vann, K.R, Kutateladze, T.G. | | 登録日 | 2018-09-19 | | 公開日 | 2018-11-14 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural insights into the pi-pi-pi stacking mechanism and DNA-binding activity of the YEATS domain.
Nat Commun, 9, 2018
|
|
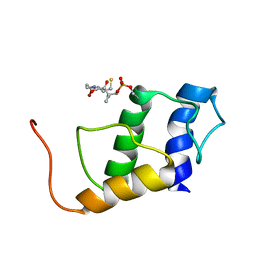
5JDX
 
 | |
2ROW
 
 | |
2TN4
 
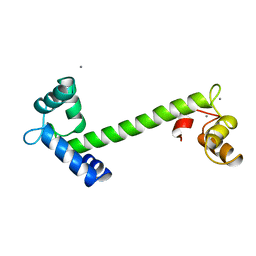 | | FOUR CALCIUM TNC | | 分子名称: | CALCIUM ION, TROPONIN C | | 著者 | Love, M.L, Dominguez, R, Houdusse, A, Cohen, C. | | 登録日 | 1997-09-18 | | 公開日 | 1998-04-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structures of four Ca2+-bound troponin C at 2.0 A resolution: further insights into the Ca2+-switch in the calmodulin superfamily.
Structure, 5, 1997
|
|
7BYU
 
 | | Crystal structure of Acidovorax avenae L-fucose mutarotase (apo form) | | 分子名称: | 1,2-ETHANEDIOL, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, L-fucose mutarotase | | 著者 | Watanabe, Y, Fukui, Y, Watanabe, S. | | 登録日 | 2020-04-24 | | 公開日 | 2020-05-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.206 Å) | | 主引用文献 | Functional and structural characterization of a novel L-fucose mutarotase involved in non-phosphorylative pathway of L-fucose metabolism.
Biochem.Biophys.Res.Commun., 528, 2020
|
|
4CBN
 
 | |
4BP0
 
 | | Crystal structure of the closed form of Pseudomonas aeruginosa SPM-1 | | 分子名称: | CHLORIDE ION, GLYCEROL, METALLO-B-LACTAMASE, ... | | 著者 | McDonough, M.A, Brem, J, Schofield, C.J. | | 登録日 | 2013-05-22 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Studying the active-site loop movement of the Sao Paolo metallo-beta-lactamase-1
Chem Sci, 6, 2015
|
|
6MV5
 
 | |
6MHI
 
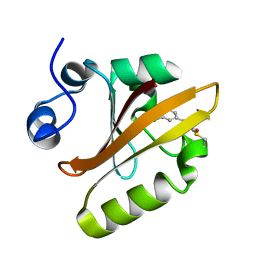 | | Photoactive Yellow Protein with covalently bound 3,5-dichloro-4-hydroxycinnamic acid chromophore | | 分子名称: | (2E)-3-(3,5-dichloro-4-hydroxyphenyl)prop-2-enoic acid, Photoactive yellow protein | | 著者 | Thomson, B.D, Both, J, Wu, Y, Parrish, R.M, Martinez, T, Boxer, S.G. | | 登録日 | 2018-09-18 | | 公開日 | 2019-05-29 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Perturbation of Short Hydrogen Bonds in Photoactive Yellow Protein via Noncanonical Amino Acid Incorporation.
J.Phys.Chem.B, 123, 2019
|
|
6MIP
 
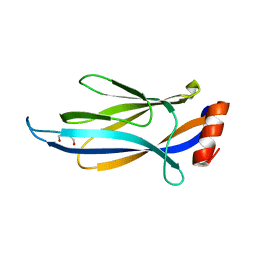 | |
7BV4
 
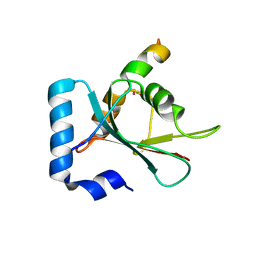 | | Crystal structure of STX17 LIR region in complex with GABARAP | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Gamma-aminobutyric acid receptor-associated protein, ... | | 著者 | Li, Y, Pan, L.F. | | 登録日 | 2020-04-09 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Decoding three distinct states of the Syntaxin17 SNARE motif in mediating autophagosome-lysosome fusion.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4EO0
 
 | | crystal structure of the pilus binding domain of the filamentous phage IKe | | 分子名称: | Attachment protein G3P | | 著者 | Jakob, R.P, Geitner, A.J, Weininger, U, Balbach, J, Dobbek, H, Schmid, F.X. | | 登録日 | 2012-04-13 | | 公開日 | 2012-05-30 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Structural and energetic basis of infection by the filamentous bacteriophage IKe.
Mol.Microbiol., 84, 2012
|
|
4ERD
 
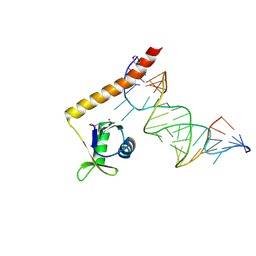 | | Crystal structure of the C-terminal domain of Tetrahymena telomerase protein p65 in complex with stem IV of telomerase RNA | | 分子名称: | 5'-R(P*GP*GP*UP*CP*GP*AP*CP*AP*UP*CP*UP*UP*CP*GP*GP*AP*UP*GP*GP*AP*CP*C)-3', POTASSIUM ION, Telomerase associated protein p65 | | 著者 | Singh, M, Wang, Z, Koo, B.-K, Patel, A, Cascio, D, Collins, K, Feigon, J. | | 登録日 | 2012-04-19 | | 公開日 | 2012-06-20 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.589 Å) | | 主引用文献 | Structural Basis for Telomerase RNA Recognition and RNP Assembly by the Holoenzyme La Family Protein p65.
Mol.Cell, 47, 2012
|
|
4EVU
 
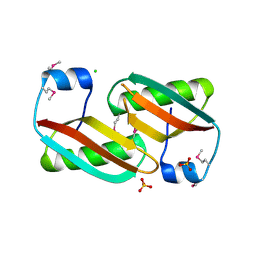 | | Crystal structure of C-terminal domain of putative periplasmic protein ydgH from S. enterica | | 分子名称: | CHLORIDE ION, Putative periplasmic protein ydgH, SULFATE ION | | 著者 | Michalska, K, Cui, H, Xu, X, Brown, R.N, Cort, J.R, Heffron, F, Nakayasu, E.S, Savchenko, A, Adkins, J.N, Joachimiak, A, Program for the Characterization of Secreted Effector Proteins (PCSEP), Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2012-04-26 | | 公開日 | 2012-05-30 | | 最終更新日 | 2014-07-30 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structural and Functional Characterization of DUF1471 Domains of Salmonella Proteins SrfN, YdgH/SssB, and YahO.
Plos One, 9, 2014
|
|
4E92
 
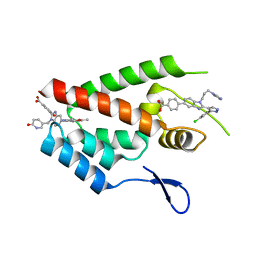 | | Crystal Structure of the N-Terminal Domain of HIV-1 Capsid in Complex With Inhibitor BM4 | | 分子名称: | 3-{5-[3-ethyl-5-(5-methylfuran-2-yl)-1H-pyrazol-1-yl]-1-[(6-oxo-1,6-dihydropyridin-3-yl)methyl]-1H-benzimidazol-2-yl}-4-hydroxybenzoic acid, 4-{2-[5-(3-chlorophenyl)-1H-pyrazol-4-yl]-1-[3-(1H-imidazol-1-yl)propyl]-1H-benzimidazol-5-yl}benzoic acid, Gag protein | | 著者 | Lemke, C.T. | | 登録日 | 2012-03-20 | | 公開日 | 2012-04-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Distinct Effects of Two HIV-1 Capsid Assembly Inhibitor Families That Bind the Same Site within the N-Terminal Domain of the Viral CA Protein.
J.Virol., 86, 2012
|
|
4E98
 
 | |
