3GQG
 
 | | Crystal structure at acidic pH of the ferric form of the Root effect hemoglobin from Trematomus bernacchii. | | 分子名称: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Vergara, A, Franzese, M, Merlino, A, Bonomi, G, Mazzarella, L. | | 登録日 | 2009-03-24 | | 公開日 | 2009-10-13 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Correlation between hemichrome stability and the root effect in tetrameric hemoglobins.
Biophys.J., 97, 2009
|
|
4TZ9
 
 | | Structure of Metallo-beta-lactamase | | 分子名称: | CADMIUM ION, COBALT (II) ION, Class B metallo-beta-lactamase, ... | | 著者 | Ferguson, J.A, Brem, J, Makena, A, McDonough, A.M, Schofield, C.J. | | 登録日 | 2014-07-09 | | 公開日 | 2015-07-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.1269 Å) | | 主引用文献 | Structure of Metallo-beta-lactamase
To Be Published
|
|
4TZE
 
 | | Structure of metallo-beta-lactamase | | 分子名称: | Class B carbapenemase NDM-5, ZINC ION | | 著者 | Ferguson, J.A, Makena, A, Brem, J, McDonough, M.A, Schofield, C.J. | | 登録日 | 2014-07-10 | | 公開日 | 2015-07-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.574 Å) | | 主引用文献 | stucuture of metallo-beta-lactamase
To Be Published
|
|
4U1D
 
 | |
1DZZ
 
 | |
1DYN
 
 | | CRYSTAL STRUCTURE AT 2.2 ANGSTROMS RESOLUTION OF THE PLECKSTRIN HOMOLOGY DOMAIN FROM HUMAN DYNAMIN | | 分子名称: | DYNAMIN | | 著者 | Ferguson, K.M, Lemmon, M.A, Schlessinger, J, Sigler, P.B. | | 登録日 | 1994-12-21 | | 公開日 | 1995-02-27 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure at 2.2 A resolution of the pleckstrin homology domain from human dynamin.
Cell(Cambridge,Mass.), 79, 1994
|
|
3HD0
 
 | | Crystal structure of Tm1865, an Endonuclease V from Thermotoga Maritima | | 分子名称: | Endonuclease V | | 著者 | Utepbergenov, D, Cooper, D.R, Derewenda, U, Derewenda, Z.S, Integrated Center for Structure and Function Innovation (ISFI) | | 登録日 | 2009-05-06 | | 公開日 | 2009-07-14 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of Tm1865, an Endonuclease V from Thermotoga Maritima
To be Published
|
|
1EGI
 
 | | STRUCTURE OF A C-TYPE CARBOHYDRATE-RECOGNITION DOMAIN (CRD-4) FROM THE MACROPHAGE MANNOSE RECEPTOR | | 分子名称: | CALCIUM ION, MACROPHAGE MANNOSE RECEPTOR | | 著者 | Feinberg, H, Park-Snyder, S, Kolatkar, A.R, Heise, C.T, Taylor, M.E, Weis, W.I. | | 登録日 | 2000-02-15 | | 公開日 | 2000-08-30 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of a C-type carbohydrate recognition domain from the macrophage mannose receptor.
J.Biol.Chem., 275, 2000
|
|
6NWZ
 
 | | Crystal structure of Agd3 a novel carbohydrate deacetylase | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | 著者 | Bamford, N.C, Howell, P.L. | | 登録日 | 2019-02-07 | | 公開日 | 2020-02-12 | | 最終更新日 | 2020-08-26 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural and biochemical characterization of the exopolysaccharide deacetylase Agd3 required for Aspergillus fumigatus biofilm formation.
Nat Commun, 11, 2020
|
|
7AA2
 
 | | Chaetomium thermophilum FAD-dependent oxidoreductase in complex with ABTS | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-ETHYL-2-[(2Z)-2-(3-ETHYL-6-SULFO-1,3-BENZOTHIAZOL-2(3H)-YLIDENE)HYDRAZINO]-6-SULFO-3H-1,3-BENZOTHIAZOL-1-IUM, ... | | 著者 | Svecova, L, Skalova, T, Kolenko, P, Koval, T, Oestergaard, L.H, Dohnalek, J. | | 登録日 | 2020-09-03 | | 公開日 | 2021-05-26 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystallographic fragment screening-based study of a novel FAD-dependent oxidoreductase from Chaetomium thermophilum.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7SF3
 
 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML1006m | | 分子名称: | (1R,2S,5S)-N-{(2S,3R)-3-hydroxy-4-(methylamino)-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase, CHLORIDE ION | | 著者 | Westberg, M, Fernandez, D, Lin, M.Z. | | 登録日 | 2021-10-02 | | 公開日 | 2022-10-05 | | 最終更新日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
7SFH
 
 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML102 | | 分子名称: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-(3-phenylpropanoyl)-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase, CALCIUM ION | | 著者 | Westberg, M, Fernandez, D, Lin, M.Z. | | 登録日 | 2021-10-03 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Rational design of a new class of protease inhibitors for the potential treatment of coronavirus diseases
To Be Published
|
|
7SFB
 
 | |
7SFI
 
 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML104 | | 分子名称: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[N-(2,4,6-trifluorophenyl)glycyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase, CALCIUM ION, ... | | 著者 | Westberg, M, Fernandez, D, Lin, M.Z. | | 登録日 | 2021-10-03 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Rational design of a new class of protease inhibitors for the potential treatment of coronavirus diseases
To Be Published
|
|
7SET
 
 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML1000 | | 分子名称: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase, CHLORIDE ION | | 著者 | Westberg, M, Fernandez, D, Lin, M.Z. | | 登録日 | 2021-10-01 | | 公開日 | 2022-10-05 | | 最終更新日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
7TAA
 
 | |
6ET9
 
 | | Structure of the acetoacetyl-CoA-thiolase/HMG-CoA-synthase complex from Methanothermococcus thermolithotrophicus at 2.75 A | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Acetyl-CoA acetyltransferase thiolase, CHLORIDE ION, ... | | 著者 | Engilberge, S, Voegeli, B, Girard, E, Riobe, F, Maury, O, Erb, T.J, Shima, S, Wagner, T. | | 登録日 | 2017-10-25 | | 公開日 | 2018-03-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Archaeal acetoacetyl-CoA thiolase/HMG-CoA synthase complex channels the intermediate via a fused CoA-binding site.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1YFD
 
 | | Crystal structure of the Y122H mutant of ribonucleotide reductase R2 protein from E. coli | | 分子名称: | MERCURY (II) ION, MU-OXO-DIIRON, Ribonucleoside-diphosphate reductase 1 beta chain | | 著者 | Kolberg, M, Logan, D.T, Bleifuss, G, Poetsch, S, Sjoeberg, B.M, Graeslund, A, Lubitz, W, Lassmann, G, Lendzian, F. | | 登録日 | 2004-12-31 | | 公開日 | 2005-02-15 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A new tyrosyl radical on Phe208 as ligand to the diiron center in Escherichia coli ribonucleotide reductase, mutant R2-Y122H. Combined x-ray diffraction and EPR/ENDOR studies
J.Biol.Chem., 280, 2005
|
|
1Y55
 
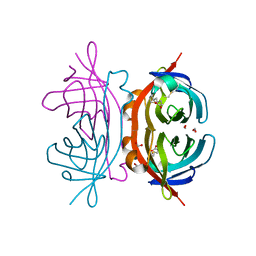 | | Crystal structure of the C122S mutant of E. Coli expressed avidin related protein 4 (AVR4)-biotin complex | | 分子名称: | Avidin-related protein 4/5, BIOTIN, FORMIC ACID | | 著者 | Eisenberg-Domovich, Y, Hytonen, V.P, Wilchek, M, Bayer, E.A, Kulomaa, M.S, Livnah, O. | | 登録日 | 2004-12-02 | | 公開日 | 2005-05-24 | | 最終更新日 | 2021-11-10 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | High-resolution crystal structure of an avidin-related protein: insight into high-affinity biotin binding and protein stability.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
6W2Y
 
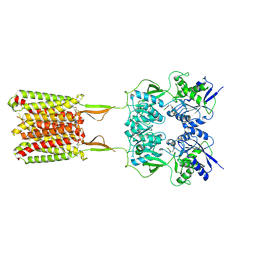 | | CryoEM Structure of GABAB1b Homodimer | | 分子名称: | (1S)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Papasergi-Scott, M.M, Robertson, M.J, Skiniotis, G. | | 登録日 | 2020-03-08 | | 公開日 | 2020-07-01 | | 最終更新日 | 2020-08-26 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structures of metabotropic GABABreceptor.
Nature, 584, 2020
|
|
1Y53
 
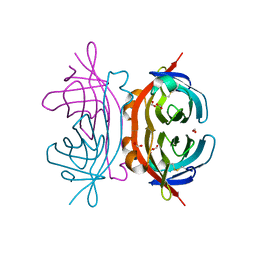 | | Crystal structure of bacterial expressed avidin related protein 4 (AVR4) C122S | | 分子名称: | Avidin-related protein 4/5, FORMIC ACID | | 著者 | Eisenberg-Domovich, Y, Hytonen, V.P, Wilchek, M, Bayer, E.A, Kulomaa, M.S, Livnah, O. | | 登録日 | 2004-12-02 | | 公開日 | 2005-05-24 | | 最終更新日 | 2021-11-10 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | High-resolution crystal structure of an avidin-related protein: insight into high-affinity biotin binding and protein stability.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1Y52
 
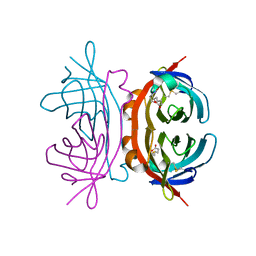 | | structure of insect cell (Baculovirus) expressed AVR4 (C122S)-biotin complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Avidin-related protein 4/5, BIOTIN | | 著者 | Eisenberg-Domovich, Y, Hytonen, V.P, Wilchek, M, Bayer, E.A, Kulomaa, M.S, Livnah, O. | | 登録日 | 2004-12-02 | | 公開日 | 2005-05-24 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | High-resolution crystal structure of an avidin-related protein: insight into high-affinity biotin binding and protein stability.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
7U92
 
 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML1006a | | 分子名称: | (1R,2S,5S)-N-{(2S,3R)-4-(azetidin-1-yl)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase, CHLORIDE ION | | 著者 | Westberg, M, Fernandez, D, Lin, M.Z. | | 登録日 | 2022-03-09 | | 公開日 | 2023-09-06 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
6QE8
 
 | | Crystal structure of Aspergillus niger GH11 endoxylanase XynA in complex with xylobiose epoxide activity based probe | | 分子名称: | (1~{R},3~{S},4~{R},5~{R})-5-[(2~{S},3~{R},4~{S},5~{R})-3,4,5-tris(oxidanyl)oxan-2-yl]oxycyclohexane-1,2,3,4-tetrol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Endo-1,4-beta-xylanase A, ... | | 著者 | Wu, L, Rowland, R.J, Davies, G.J. | | 登録日 | 2019-01-07 | | 公開日 | 2019-06-05 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Dynamic and Functional Profiling of Xylan-Degrading Enzymes inAspergillusSecretomes Using Activity-Based Probes.
Acs Cent.Sci., 5, 2019
|
|
6EU9
 
 | | Crystal structure of Platynereis dumerilii RAR ligand-binding domain in complex with all-trans retinoic acid | | 分子名称: | RETINOIC ACID, Retinoic acid receptor | | 著者 | Handberg-Thorsager, M, Gutierrez-Mazariegos, J, Arold, S.T, Nadendla, E.K, Bertucci, P.Y, Germain, P, Tomancak, P, Pierzchalski, K, Jones, J.W, Albalat, R, Kane, M.A, Bourguet, W, Laudet, V, Arendt, D, Schubert, M. | | 登録日 | 2017-10-29 | | 公開日 | 2018-03-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | The ancestral retinoic acid receptor was a low-affinity sensor triggering neuronal differentiation.
Sci Adv, 4, 2018
|
|
