3P7P
 
 | |
3P7Y
 
 | |
3P7W
 
 | |
3P94
 
 | |
3P9I
 
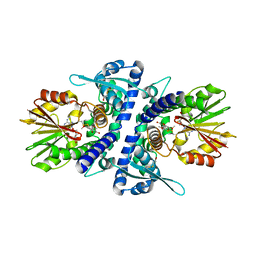 | | Crystal structure of perennial ryegrass LpOMT1 complexed with S-adenosyl-L-homocysteine and sinapaldehyde | | 分子名称: | (2E)-3-(4-hydroxy-3,5-dimethoxyphenyl)prop-2-enal, BETA-MERCAPTOETHANOL, Caffeic acid O-methyltransferase, ... | | 著者 | Louie, G.V, Noel, J.P, Bowman, M.E. | | 登録日 | 2010-10-17 | | 公開日 | 2011-01-12 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structure-Function Analyses of a Caffeic Acid O-Methyltransferase from Perennial Ryegrass Reveal the Molecular Basis for Substrate Preference.
Plant Cell, 22, 2010
|
|
3P9Q
 
 | | Structure of I274C variant of E. coli KatE | | 分子名称: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE 17R, 18S, ... | | 著者 | Loewen, P.C, Jha, V, Louis, S, Chelikani, P, Carpena, X, Fita, I. | | 登録日 | 2010-10-18 | | 公開日 | 2010-12-22 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Modulation of heme orientation and binding by a single residue in catalase HPII of Escherichia coli.
Biochemistry, 50, 2011
|
|
3PA5
 
 | |
3OKO
 
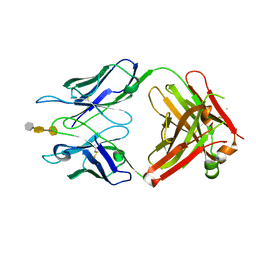 | | Crystal structure of S25-39 in complex with Kdo(2.8)Kdo(2.4)Kdo | | 分子名称: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-8)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-prop-2-en-1-yl 3-deoxy-alpha-D-manno-oct-2-ulopyranosidonic acid, S25-39 Fab (IgG1k) heavy chain, S25-39 Fab (IgG1k) light chain, ... | | 著者 | Blackler, R.J, Evans, S.V. | | 登録日 | 2010-08-25 | | 公開日 | 2011-04-06 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | A Common NH53K Mutation in the Combining Site of Antibodies Raised against Chlamydial LPS Glycoconjugates Significantly Increases Avidity.
Biochemistry, 50, 2011
|
|
3PB3
 
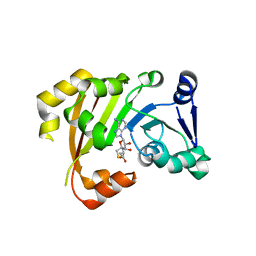 | | Structure of an Antibiotic Related Methyltransferase | | 分子名称: | 16S rRNA methylase, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Sivaraman, J, Husain, N. | | 登録日 | 2010-10-20 | | 公開日 | 2010-11-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis for the methylation of A1408 in 16S rRNA by a panaminoglycoside resistance methyltransferase NpmA from a clinical isolate and analysis of the NpmA interactions with the 30S ribosomal subunit.
Nucleic Acids Res., 2010
|
|
3OMK
 
 | |
3OMS
 
 | | Putative 3-demethylubiquinone-9 3-methyltransferase, PhnB protein, from Bacillus cereus. | | 分子名称: | 1,2-ETHANEDIOL, PhnB protein | | 著者 | Osipiuk, J, Li, H, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-08-27 | | 公開日 | 2010-09-08 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | X-ray crystal structure of putative 3-demethylubiquinone-9
3-methyltransferase, PhnB protein, from Bacillus cereus.
To be Published
|
|
3OO7
 
 | |
3P7S
 
 | |
3OPW
 
 | | Crystal Structure of the Rph1 catalytic core | | 分子名称: | DNA damage-responsive transcriptional repressor RPH1 | | 著者 | Chang, Y, Wu, J, Tong, X, Zhou, J, Ding, J. | | 登録日 | 2010-09-02 | | 公開日 | 2010-12-22 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of the catalytic core of Saccharomyces cerevesiae histone demethylase Rph1: insights into the substrate specificity and catalytic mechanism
Biochem.J., 433, 2011
|
|
3P81
 
 | |
3OQF
 
 | |
3P9S
 
 | | Structure of I274A variant of E. coli KatE | | 分子名称: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE 17R, 18S, ... | | 著者 | Loewen, P.C, Jha, V, Louis, S, Chelikani, P, Carpena, X, Fita, I. | | 登録日 | 2010-10-18 | | 公開日 | 2010-12-22 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Modulation of heme orientation and binding by a single residue in catalase HPII of Escherichia coli.
Biochemistry, 50, 2011
|
|
3OSO
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS L25A at cryogenic temperature | | 分子名称: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | 著者 | Caro, J.A, Schlessman, J.L, Garcia-Moreno, E.B, Heroux, A. | | 登録日 | 2010-09-09 | | 公開日 | 2011-09-21 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Cavities determine the pressure unfolding of proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3OOE
 
 | | Crystal structure of E. Coli purine nucleoside phosphorylase with PO4 | | 分子名称: | PHOSPHATE ION, Purine nucleoside phosphorylase deoD-type | | 著者 | Mikleusevic, G, Stefanic, Z, Narzyk, M, Wielgus-Kutrowska, B, Bzowska, A, Luic, M. | | 登録日 | 2010-08-31 | | 公開日 | 2011-07-13 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Validation of the catalytic mechanism of Escherichia coli purine nucleoside phosphorylase by structural and kinetic studies.
Biochimie, 93, 2011
|
|
3OPT
 
 | | Crystal structure of the Rph1 catalytic core with a-ketoglutarate | | 分子名称: | 2-OXOGLUTARIC ACID, DNA damage-responsive transcriptional repressor RPH1, NICKEL (II) ION | | 著者 | Chang, Y, Wu, J, Tong, X, Zhou, J, Ding, J. | | 登録日 | 2010-09-02 | | 公開日 | 2010-12-22 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of the catalytic core of Saccharomyces cerevesiae histone demethylase Rph1: insights into the substrate specificity and catalytic mechanism
Biochem.J., 433, 2011
|
|
3OV2
 
 | | Curcumin synthase 1 from Curcuma longa | | 分子名称: | 1,2-ETHANEDIOL, Curcumin synthase, MALONATE ION | | 著者 | Katsuyama, Y, Miyazono, K, Tanokura, M, Ohnishi, Y, Horinouchi, S. | | 登録日 | 2010-09-15 | | 公開日 | 2010-12-08 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | A hydrophobic cavity discovered in a curcumin synthase facilitates utilization of a beta-keto acid as an extender substrate for the atypical type III polyleteide synthase
To be Published
|
|
3OW7
 
 | |
3OQ6
 
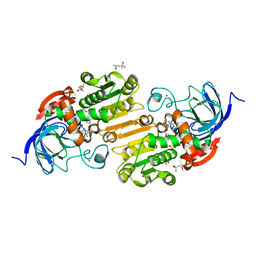 | | Horse liver alcohol dehydrogenase A317C mutant complexed with NAD+ and 2,3,4,5,6-pentafluorobenzyl alcohol | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2,3,4,5,6-PENTAFLUOROBENZYL ALCOHOL, Alcohol dehydrogenase E chain, ... | | 著者 | Plapp, B.V, Herdendorf, T.J. | | 登録日 | 2010-09-02 | | 公開日 | 2010-11-10 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Origins of the high catalytic activity of human alcohol dehydrogenase 4 studied with horse liver A317C alcohol dehydrogenase.
Chem.Biol.Interact, 191, 2011
|
|
3OXG
 
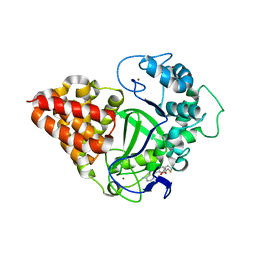 | | human lysine methyltransferase Smyd3 in complex with AdoHcy (Form III) | | 分子名称: | S-ADENOSYL-L-HOMOCYSTEINE, SET and MYND domain-containing protein 3, ZINC ION | | 著者 | Xu, S, Wu, J, Sun, B, Zhong, C, Ding, J. | | 登録日 | 2010-09-21 | | 公開日 | 2011-02-23 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.41 Å) | | 主引用文献 | Structural and biochemical studies of human lysine methyltransferase Smyd3 reveal the important functional roles of its post-SET and TPR domains and the regulation of its activity by DNA binding.
Nucleic Acids Res., 39, 2011
|
|
3OYE
 
 | |
