3PBJ
 
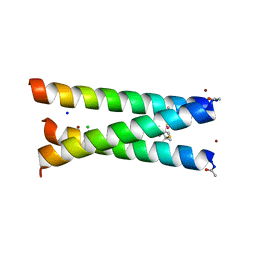 | | Hydrolytic catalysis and structural stabilization in a designed metalloprotein | | 分子名称: | CHLORIDE ION, COIL SER L9L-Pen L23H, MERCURY (II) ION, ... | | 著者 | Zastrow, M.L, Peacock, A.F.A, Stuckey, J.A, Pecoraro, V.L. | | 登録日 | 2010-10-20 | | 公開日 | 2011-11-30 | | 最終更新日 | 2022-05-04 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Hydrolytic catalysis and structural stabilization in a designed metalloprotein.
Nat Chem, 4, 2012
|
|
3U0S
 
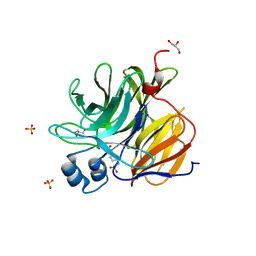 | | Crystal Structure of an Enzyme Redesigned Through Multiplayer Online Gaming: CE6 | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Diisopropyl-fluorophosphatase, GLYCEROL, ... | | 著者 | Bale, J.B, Shen, B.W, Stoddard, B.L. | | 登録日 | 2011-09-29 | | 公開日 | 2012-02-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Increased Diels-Alderase activity through backbone remodeling guided by Foldit players.
Nat.Biotechnol., 30, 2012
|
|
3TWG
 
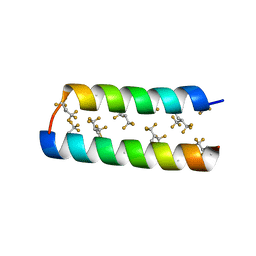 | |
3TWF
 
 | | Crystal structure of the de novo designed fluorinated peptide alpha4F3a | | 分子名称: | ACETYL GROUP, SODIUM ION, TRIETHYLENE GLYCOL, ... | | 著者 | Buer, B.C, Meagher, J.L, Stuckey, J.A, Marsh, E.N.G. | | 登録日 | 2011-09-21 | | 公開日 | 2012-03-14 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Structural basis for the enhanced stability of highly fluorinated proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6REN
 
 | | Crystal structure of 3fPizza6-SH with Zn2+ | | 分子名称: | 3fPizza6-SH, GLYCEROL, ISOPROPYL ALCOHOL, ... | | 著者 | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | 登録日 | 2019-04-12 | | 公開日 | 2019-07-31 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Artificial beta-propeller protein-based hydrolases.
Chem.Commun.(Camb.), 55, 2019
|
|
6REH
 
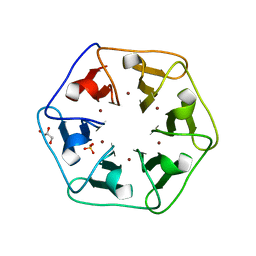 | | Crystal structure of Pizza6-S with Cu2+ | | 分子名称: | COPPER (II) ION, GLYCEROL, Pizza6-S, ... | | 著者 | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | 登録日 | 2019-04-12 | | 公開日 | 2019-07-31 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Artificial beta-propeller protein-based hydrolases.
Chem.Commun.(Camb.), 55, 2019
|
|
6REJ
 
 | | Crystal structure of Pizza6-SH with Zn2+ | | 分子名称: | Pizza6-SH, SULFATE ION, ZINC ION | | 著者 | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | 登録日 | 2019-04-12 | | 公開日 | 2019-07-31 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Artificial beta-propeller protein-based hydrolases.
Chem.Commun.(Camb.), 55, 2019
|
|
5MKE
 
 | | cryoEM Structure of Polycystin-2 in complex with cations and lipids | | 分子名称: | 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Wilkes, M, Madej, M.G, Ziegler, C. | | 登録日 | 2016-12-04 | | 公開日 | 2017-01-18 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Molecular insights into lipid-assisted Ca(2+) regulation of the TRP channel Polycystin-2.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5MKF
 
 | | cryoEM Structure of Polycystin-2 in complex with calcium and lipids | | 分子名称: | 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Wilkes, M, Madej, M.G, Ziegler, C. | | 登録日 | 2016-12-04 | | 公開日 | 2017-01-18 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Molecular insights into lipid-assisted Ca(2+) regulation of the TRP channel Polycystin-2.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6REG
 
 | | Crystal structure of Pizza6-S with Zn2+ | | 分子名称: | GLYCEROL, Pizza6-S, ZINC ION | | 著者 | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | 登録日 | 2019-04-12 | | 公開日 | 2019-07-31 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Artificial beta-propeller protein-based hydrolases.
Chem.Commun.(Camb.), 55, 2019
|
|
6REK
 
 | | Crystal structure of Pizza6-SH with Cu2+ | | 分子名称: | COPPER (II) ION, GLYCEROL, Pizza6-SH | | 著者 | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | 登録日 | 2019-04-12 | | 公開日 | 2019-07-31 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Artificial beta-propeller protein-based hydrolases.
Chem.Commun.(Camb.), 55, 2019
|
|
1E7Q
 
 | | GDP 4-keto-6-deoxy-D-mannose epimerase reductase S107A | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETYLPHOSPHATE, GDP-FUCOSE SYNTHETASE, ... | | 著者 | Rosano, C, Izzo, G, Bolognesi, M. | | 登録日 | 2000-09-07 | | 公開日 | 2000-10-18 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Probing the Catalytic Mechanism of Gdp-4-Keto-6-Deoxy-D-Mannose Epimerase/Reductase by Kinetic and Crystallographic Characterization of Site-Specific Mutants
J.Mol.Biol., 303, 2000
|
|
1E6U
 
 | | GDP 4-keto-6-deoxy-D-mannose epimerase reductase | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETYLPHOSPHATE, GDP-FUCOSE SYNTHETASE, ... | | 著者 | Rosano, C, Izzo, G, Bolognesi, M. | | 登録日 | 2000-08-23 | | 公開日 | 2000-10-22 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Probing the Catalytic Mechanism of Gdp-4-Keto-6-Deoxy-D-Mannose Epimerase/Reductase by Kinetic and Crystallographic Characterization of Site-Specific Mutants
J.Mol.Biol., 303, 2000
|
|
1E7S
 
 | | GDP 4-keto-6-deoxy-D-mannose epimerase reductase K140R | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETYLPHOSPHATE, GDP-FUCOSE SYNTHETASE, ... | | 著者 | Rosano, C, Zuccotti, S, Izzo, G, Bolognesi, M. | | 登録日 | 2000-09-07 | | 公開日 | 2000-10-18 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Probing the Catalytic Mechanism of Gdp-4-Keto-6-Deoxy-D-Mannose Epimerase/Reductase by Kinetic and Crystallographic Characterization of Site-Specific Mutants
J.Mol.Biol., 303, 2000
|
|
1E7R
 
 | | GDP 4-keto-6-deoxy-D-mannose epimerase reductase Y136E | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETYLPHOSPHATE, GDP-FUCOSE SYNTHETASE, ... | | 著者 | Rosano, C, Izzo, G, Bolognesi, M. | | 登録日 | 2000-09-07 | | 公開日 | 2000-10-18 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Probing the Catalytic Mechanism of Gdp-4-Keto-6-Deoxy-D-Mannose Epimerase/Reductase by Kinetic and Crystallographic Characterization of Site-Specific Mutants
J.Mol.Biol., 303, 2000
|
|
5KAF
 
 | | RT XFEL structure of Photosystem II in the dark state at 3.0 A resolution | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | 登録日 | 2016-06-01 | | 公開日 | 2016-11-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.00001 Å) | | 主引用文献 | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
5KAI
 
 | | NH3-bound RT XFEL structure of Photosystem II 500 ms after the 2nd illumination (2F) at 2.8 A resolution | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | 登録日 | 2016-06-01 | | 公開日 | 2016-11-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.80000925 Å) | | 主引用文献 | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
2JOF
 
 | |
2N35
 
 | |
2N8D
 
 | |
7B1X
 
 | | Crystal structure of cold-active esterase PMGL3 from permafrost metagenomic library | | 分子名称: | esterase PMGL3 | | 著者 | Boyko, K.M, Nikolaeva, A.Y, Petrovskaya, L.E, Kryukova, M.V, Kryukova, E.A, Korzhenevsky, D.A, Lomakina, G.Y, Novototskaya-Vlasova, K.A, Rivkina, E.M, Dolgikh, D.A, Kirpichnikov, M.P, Popov, V.O. | | 登録日 | 2020-11-25 | | 公開日 | 2021-11-03 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural and Biochemical Characterization of a Cold-Active PMGL3 Esterase with Unusual Oligomeric Structure.
Biomolecules, 11, 2021
|
|
7L85
 
 | |
1P9B
 
 | | Structure of fully ligated Adenylosuccinate synthetase from Plasmodium falciparum | | 分子名称: | 6-O-PHOSPHORYL INOSINE MONOPHOSPHATE, Adenylosuccinate Synthetase, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Eaazhisai, K, Jayalakshmi, R, Gayathri, P, Anand, R.P, Sumathy, K, Balaram, H, Murthy, M.R. | | 登録日 | 2003-05-10 | | 公開日 | 2004-02-03 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of Fully Ligated Adenylosuccinate Synthetase from Plasmodium falciparum.
J.Mol.Biol., 335, 2004
|
|
3PFD
 
 | |
4N6U
 
 | | Adhiron: a stable and versatile peptide display scaffold - truncated adhiron | | 分子名称: | Adhiron | | 著者 | Mcpherson, M, Tomlinson, D, Owen, R.L, Nettleship, J.E, Owens, R.J. | | 登録日 | 2013-10-14 | | 公開日 | 2014-04-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.251 Å) | | 主引用文献 | Adhiron: a stable and versatile peptide display scaffold for molecular recognition applications.
Protein Eng.Des.Sel., 27, 2014
|
|
