1GMQ
 
 | | COMPLEX OF RIBONUCLEASE FROM STREPTOMYCES AUREOFACIENS WITH 2'-GMP AT 1.7 ANGSTROMS RESOLUTION | | 分子名称: | RIBONUCLEASE SA, SULFATE ION | | 著者 | Sevcik, J, Hill, C, Dauter, Z, Wilson, K. | | 登録日 | 1992-10-01 | | 公開日 | 1993-10-31 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Complex of ribonuclease from Streptomyces aureofaciens with 2'-GMP at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
4NJV
 
 | | Crystal structure of multidrug-resistant clinical isolate A02 HIV-1 protease in complex with ritonavir | | 分子名称: | Protease, RITONAVIR | | 著者 | Yedidi, R.S, Garimella, H, Chang, S.B, Kaufman, J.D, Das, D, Wingfield, P.T, Mitsuya, H. | | 登録日 | 2013-11-11 | | 公開日 | 2014-04-02 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A Conserved Hydrogen-Bonding Network of P2 bis-Tetrahydrofuran-Containing HIV-1 Protease Inhibitors (PIs) with a Protease Active-Site Amino Acid Backbone Aids in Their Activity against PI-Resistant HIV.
Antimicrob.Agents Chemother., 58, 2014
|
|
1GNF
 
 | | SOLUTION STRUCTURE OF THE N-TERMINAL ZINC FINGER OF MURINE GATA-1, NMR, 25 STRUCTURES | | 分子名称: | TRANSCRIPTION FACTOR GATA-1, ZINC ION | | 著者 | Kowalski, K, Czolij, R, King, G.F, Crossley, M, Mackay, J.P. | | 登録日 | 1998-10-12 | | 公開日 | 1999-06-08 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of the N-terminal zinc finger of GATA-1 reveals a specific binding face for the transcriptional co-factor FOG.
J.Biomol.NMR, 13, 1999
|
|
3AEO
 
 | | Reaction intermediate structure of Entamoeba histolytica methionine gamma-lyase 1 containing methionine alpha, beta-enamine-pyridoxamine-5'-phosphate | | 分子名称: | (2E)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]-4-(methylsulfanyl)but-2-enoic acid, GLYCEROL, Methionine gamma-lyase, ... | | 著者 | Karaki, T, Sato, D, Shimizu, A, Nozaki, T, Harada, S. | | 登録日 | 2010-02-10 | | 公開日 | 2011-02-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystal structure of Entamoeba histolytica methionine gamma-lyase 1
To be Published
|
|
1GMP
 
 | | COMPLEX OF RIBONUCLEASE FROM STREPTOMYCES AUREOFACIENS WITH 2'-GMP AT 1.7 ANGSTROMS RESOLUTION | | 分子名称: | GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE SA, SULFATE ION | | 著者 | Sevcik, J, Hill, C, Dauter, Z, Wilson, K. | | 登録日 | 1992-10-01 | | 公開日 | 1993-10-31 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Complex of ribonuclease from Streptomyces aureofaciens with 2'-GMP at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1GMR
 
 | | COMPLEX OF RIBONUCLEASE FROM STREPTOMYCES AUREOFACIENS WITH 2'-GMP AT 1.7 ANGSTROMS RESOLUTION | | 分子名称: | GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE SA, SULFATE ION | | 著者 | Sevcik, J, Hill, C, Dauter, Z, Wilson, K. | | 登録日 | 1992-10-01 | | 公開日 | 1993-10-31 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Complex of ribonuclease from Streptomyces aureofaciens with 2'-GMP at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
2X8F
 
 | | Native structure of Endo-1,5-alpha-L-arabinanases from Bacillus subtilis | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | 著者 | de Sanctis, D, Inacio, J.M, Lindley, P.F, de Sa-Nogueira, I, Bento, I. | | 登録日 | 2010-03-09 | | 公開日 | 2011-03-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | New Evidence for the Role of Calcium in the Glycosidase Reaction of Gh43 Arabinanases.
FEBS J., 277, 2010
|
|
1P53
 
 | | The Crystal Structure of ICAM-1 D3-D5 fragment | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Intercellular adhesion molecule-1 | | 著者 | Yang, Y, Jun, C.D, Liu, J.H, Zhang, R, Jochimiak, A, Springer, T.A, Wang, J.H. | | 登録日 | 2003-04-24 | | 公開日 | 2004-05-04 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (3.06 Å) | | 主引用文献 | Structural basis for dimerization of ICAM-1 on the cell surface.
Mol.Cell, 14, 2004
|
|
3S2C
 
 | | Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 | | 分子名称: | Alpha-N-arabinofuranosidase | | 著者 | Souza, T.A.C.B, Santos, C.R, Souza, A.R, Oldiges, D.P, Ruller, R, Prade, R.A, Squina, F.M, Murakami, M.T. | | 登録日 | 2011-05-16 | | 公開日 | 2012-02-01 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure of a novel thermostable GH51 Alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1
Protein Sci., 20, 2011
|
|
5Z56
 
 | | cryo-EM structure of a human activated spliceosome (mature Bact) at 5.1 angstrom. | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | 著者 | Zhang, X, Yan, C, Zhan, X, Li, L, Lei, J, Shi, Y. | | 登録日 | 2018-01-17 | | 公開日 | 2018-09-19 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (5.1 Å) | | 主引用文献 | Structure of the human activated spliceosome in three conformational states.
Cell Res., 28, 2018
|
|
4CNM
 
 | | Crystal structure of human 5T4 (Wnt-activated inhibitory factor 1, Trophoblast glycoprotein) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, ... | | 著者 | Zhao, Y, Malinauskas, T, Harlos, K, Jones, E.Y. | | 登録日 | 2014-01-23 | | 公開日 | 2014-02-26 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural Insights Into the Inhibition of Wnt Signaling by Cancer Antigen 5T4/Wnt-Activated Inhibitory Factor 1.
Structure, 22, 2014
|
|
2FYB
 
 | | Crystal structure of the catalytic domain of the human beta1,4-galactosyltransferase mutant M339H in complex with Mn and UDP-galactose in open conformation | | 分子名称: | Beta-1,4-galactosyltransferase 1, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE | | 著者 | Ramakrishnan, B, Ramasamy, V, Qasba, P.K. | | 登録日 | 2006-02-07 | | 公開日 | 2006-03-14 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural Snapshots of beta-1,4-Galactosyltransferase-I Along the Kinetic Pathway.
J.Mol.Biol., 357, 2006
|
|
4CS7
 
 | | Crystal structure of the asymmetric human metapneumovirus M2-1 tetramer, form 1 | | 分子名称: | M2-1, ZINC ION | | 著者 | Leyrat, C, Renner, M, Harlos, K, Grimes, J.M. | | 登録日 | 2014-03-05 | | 公開日 | 2014-05-28 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Drastic Changes in Conformational Dynamics of the Antiterminator M2-1 Regulate Transcription Efficiency in Pneumovirinae.
Elife, 3, 2014
|
|
4Q1P
 
 | |
4COP
 
 | | HIV-1 capsid C-terminal domain mutant (Y169S) | | 分子名称: | CAPSID PROTEIN P24 | | 著者 | Bharat, T.A.M, Castillo-Menendez, L.R, Hagen, W.J.H, Lux, V, Igonet, S, Schorb, M, Schur, F.K.M, Krausslich, H.-G, Briggs, J.A.G. | | 登録日 | 2014-01-29 | | 公開日 | 2014-06-04 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Cryo-Electron Microscopy of Tubular Arrays of HIV-1 Gag Resolves Structures Essential for Immature Virus Assembly.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4KBR
 
 | |
4Q1R
 
 | | Galectin-1 in Complex with Ligand AN027 | | 分子名称: | Galectin-1, SULFATE ION, propyl 2-(acetylamino)-2-deoxy-4-O-[3-O-({1-[2-(3-hydroxyphenyl)-2-oxoethyl]-1H-1,2,3-triazol-4-yl}methyl)-beta-D-galactopyranosyl]-beta-D-glucopyranoside | | 著者 | Grimm, C, Bertleff-Zieschang, N. | | 登録日 | 2014-04-04 | | 公開日 | 2015-10-07 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Galectin-1 in Complex with Ligand AN027
To be Published
|
|
1JM4
 
 | | NMR Structure of P/CAF Bromodomain in Complex with HIV-1 Tat Peptide | | 分子名称: | HIV-1 Tat Peptide, P300/CBP-associated Factor | | 著者 | Mujtaba, S, He, Y, Zeng, L, Farooq, A, Carlson, J.E, Ott, M, Verdin, E, Zhou, M.-M. | | 登録日 | 2001-07-17 | | 公開日 | 2002-07-17 | | 最終更新日 | 2024-10-30 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis of lysine-acetylated HIV-1 Tat recognition by PCAF bromodomain
Mol.Cell, 9, 2002
|
|
1PMS
 
 | | PLECKSTRIN HOMOLOGY DOMAIN OF SON OF SEVENLESS 1 (SOS1) WITH GLYCINE-SERINE ADDED TO THE N-TERMINUS, NMR, 20 STRUCTURES | | 分子名称: | SOS 1 | | 著者 | Koshiba, S, Kigawa, T, Kim, J, Shirouzu, M, Bowtell, D, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 1997-02-18 | | 公開日 | 1997-05-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of the pleckstrin homology domain of mouse Son-of-sevenless 1 (mSos1).
J.Mol.Biol., 269, 1997
|
|
1UC5
 
 | | Structure of diol dehydratase complexed with (R)-1,2-propanediol | | 分子名称: | AMMONIUM ION, CYANOCOBALAMIN, POTASSIUM ION, ... | | 著者 | Shibata, N, Nakanishi, Y, Fukuoka, M, Yamanishi, M, Yasuoka, N, Toraya, T. | | 登録日 | 2003-04-08 | | 公開日 | 2003-07-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural rationalization for the lack of stereospecificity in coenzyme B12-dependent diol dehydratase
J.Biol.Chem., 278, 2003
|
|
4Q78
 
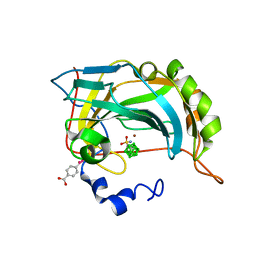 | | Structure-assisted design of carborane-based inhibitors of carbonic anhydrase | | 分子名称: | 1-(sulfamoylamino)methyl-1,2-dicarba-closo-dodecaborane, Carbonic anhydrase 2, MERCURIBENZOIC ACID, ... | | 著者 | Mader, P, Brynda, J, Rezacova, P. | | 登録日 | 2014-04-24 | | 公開日 | 2015-03-18 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Carborane-based carbonic anhydrase inhibitors: insight into CAII/CAIX specificity from a high-resolution crystal structure, modeling, and quantum chemical calculations.
Biomed Res Int, 2014, 2014
|
|
3U54
 
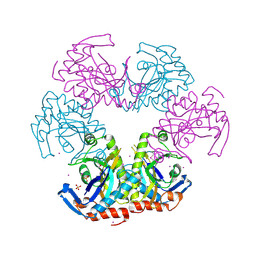 | | Crystal structure (Type-1) of SAICAR synthetase from Pyrococcus horikoshii OT3 | | 分子名称: | 1,4-BUTANEDIOL, ACETATE ION, CADMIUM ION, ... | | 著者 | Manjunath, K, Kanaujia, S.P, Kanagaraj, S, Jeyakanthan, J, Sekar, K. | | 登録日 | 2011-10-11 | | 公開日 | 2012-10-17 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structure of SAICAR synthetase from Pyrococcus horikoshii OT3: insights into thermal stability
Int.J.Biol.Macromol., 53, 2013
|
|
7L21
 
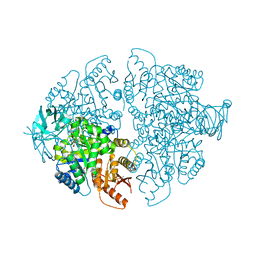 | | Pyruvate Kinase M2 mutant-N70D | | 分子名称: | 1,6-di-O-phosphono-beta-D-fructofuranose, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Nandi, S, Dey, M. | | 登録日 | 2020-12-16 | | 公開日 | 2021-12-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Identification of residues involved in allosteric signal transmission from amino acid binding site of pyruvate kinase muscle isoform 2.
Plos One, 18, 2023
|
|
5UPN
 
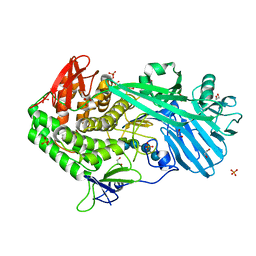 | |
2RR0
 
 | | Structure of epidermal growth factor-like repeat 12 of mouse Notch-1 receptor | | 分子名称: | Neurogenic locus notch homolog protein 1 | | 著者 | Hosoguchi, K, Shimizu, K, Fujitani, N, Nishimura, S. | | 登録日 | 2010-02-26 | | 公開日 | 2010-10-13 | | 最終更新日 | 2024-10-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Chemical Synthesis, Folding, and Structural Insights into O-Fucosylated Epidermal Growth Factor-like Repeat 12 of Mouse Notch-1 Receptor
J.Am.Chem.Soc., 132, 2010
|
|
