7WJK
 
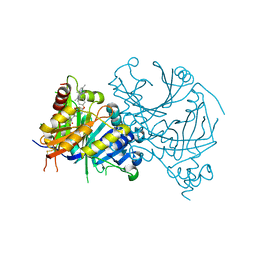 | | Complex structure of AtHPPD-ZWJ | | 分子名称: | 3-[2-[4-[[4-[2,2-bis(fluoranyl)-4,6,10,12-tetramethyl-1,3-diaza-2$l^{4}-boratricyclo[7.3.0.0^{3,7}]dodeca-4,6,9,11-tetraen-8-yl]phenoxy]methyl]-1,2,3-triazol-1-yl]ethyl]-1,5-dimethyl-6-[(1S)-2-oxidanyl-6-oxidanylidene-cyclohex-2-en-1-yl]carbonyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | 著者 | Yang, G.-F, Lin, H.-Y, Dong, J. | | 登録日 | 2022-01-06 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.704 Å) | | 主引用文献 | Complex structure of AtHPPD-PyQ1
To Be Published
|
|
1CF9
 
 | |
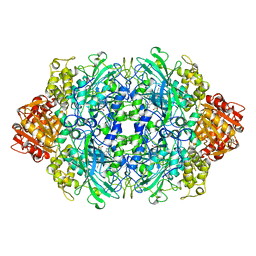
1QF7
 
 | |
3V2W
 
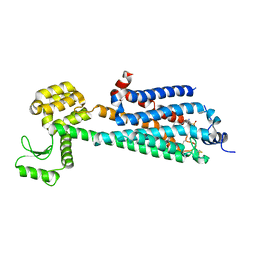 | | Crystal Structure of a Lipid G protein-Coupled Receptor at 3.35A | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Sphingosine 1-phosphate receptor 1, Lysozyme chimera, ... | | 著者 | Hanson, M.A, Roth, C.B, Jo, E, Griffith, M.T, Scott, F.L, Reinhart, G, Desale, H, Clemons, B, Cahalan, S.M, Schuerer, S.C, Sanna, M.G, Han, G.W, Kuhn, P, Rosen, H, Stevens, R.C, GPCR Network (GPCR) | | 登録日 | 2011-12-12 | | 公開日 | 2012-02-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.35 Å) | | 主引用文献 | Crystal structure of a lipid G protein-coupled receptor.
Science, 335, 2012
|
|
4W55
 
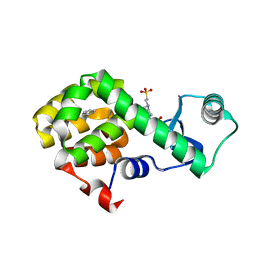 | | T4 Lysozyme L99A with n-Propylbenzene Bound | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Endolysin, propylbenzene | | 著者 | Merski, M, Shoichet, B.K, Eidam, O, Fischer, M. | | 登録日 | 2014-08-16 | | 公開日 | 2015-04-01 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.6401 Å) | | 主引用文献 | Homologous ligands accommodated by discrete conformations of a buried cavity.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4W58
 
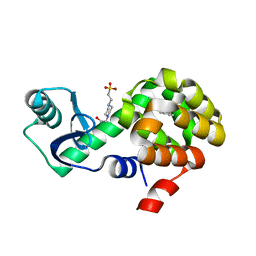 | | T4 Lysozyme L99A with n-Pentylbenzene Bound | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Endolysin, pentylbenzene | | 著者 | Merski, M, Shoichet, B.K, Eidam, O, Fischer, M. | | 登録日 | 2014-08-16 | | 公開日 | 2015-04-01 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Homologous ligands accommodated by discrete conformations of a buried cavity.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5VEW
 
 | | Structure of the human GLP-1 receptor complex with PF-06372222 | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Glucagon-like peptide 1 receptor,Endolysin chimera, N-{4-[(R)-(3,3-dimethylcyclobutyl)({6-[4-(trifluoromethyl)-1H-imidazol-1-yl]pyridin-3-yl}amino)methyl]benzene-1-carbonyl}-beta-alanine, ... | | 著者 | Song, G, Yang, D, Wang, Y, Graaf, C.D, Zhou, Q, Jiang, S, Liu, K, Cai, X, Dai, A, Lin, G, Liu, D, Wu, F, Wu, Y, Zhao, S, Ye, L, Han, G.W, Lau, J, Wu, B, Hanson, M.A, Liu, Z.-J, Wang, M.-W, Stevens, R.C. | | 登録日 | 2017-04-05 | | 公開日 | 2017-05-24 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators.
Nature, 546, 2017
|
|
5VEX
 
 | | Structure of the human GLP-1 receptor complex with NNC0640 | | 分子名称: | 4-{[(4-cyclohexylphenyl){[3-(methylsulfonyl)phenyl]carbamoyl}amino]methyl}-N-(1H-tetrazol-5-yl)benzamide, Glucagon-like peptide 1 receptor, Endolysin chimera | | 著者 | Song, G, Yang, D, Wang, Y, Graaf, C.D, Zhou, Q, Jiang, S, Liu, K, Cai, X, Dai, A, Lin, G, Liu, D, Wu, F, Wu, Y, Zhao, S, Ye, L, Han, G.W, Lau, J, Wu, B, Hanson, M.A, Liu, Z.-J, Wang, M.-W, Stevens, R.C. | | 登録日 | 2017-04-05 | | 公開日 | 2017-05-17 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators.
Nature, 546, 2017
|
|
6KK1
 
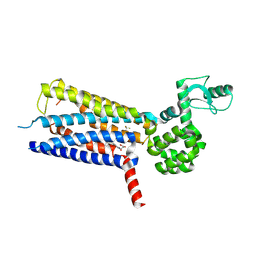 | | Structure of thermal-stabilised(M8) human GLP-1 receptor transmembrane domain | | 分子名称: | Glucagon-like peptide 1 receptor,Endolysin,Glucagon-like peptide 1 receptor, N-{4-[(R)-(3,3-dimethylcyclobutyl)({6-[4-(trifluoromethyl)-1H-imidazol-1-yl]pyridin-3-yl}amino)methyl]benzene-1-carbonyl}-beta-alanine | | 著者 | Song, G. | | 登録日 | 2019-07-23 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Mutagenesis facilitated crystallization of GLP-1R.
Iucrj, 6, 2019
|
|
6KJV
 
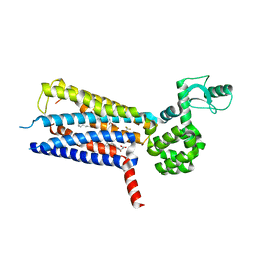 | | Structure of thermal-stabilised(M9) human GLP-1 receptor transmembrane domain | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Glucagon-like peptide 1 receptor,Endolysin,Glucagon-like peptide 1 receptor, N-{4-[(R)-(3,3-dimethylcyclobutyl)({6-[4-(trifluoromethyl)-1H-imidazol-1-yl]pyridin-3-yl}amino)methyl]benzene-1-carbonyl}-beta-alanine | | 著者 | Song, G. | | 登録日 | 2019-07-23 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Mutagenesis facilitated crystallization of GLP-1R.
Iucrj, 6, 2019
|
|
5JWS
 
 | | T4 Lysozyme L99A with 1-Hydro-2-ethyl-1,2-azaborine Bound | | 分子名称: | 2-ethyl-1,2-dihydro-1,2-azaborinine, CHLORIDE ION, Endolysin | | 著者 | Lee, H, Fischer, M, Shoichet, B.K, Liu, S.-Y. | | 登録日 | 2016-05-12 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Hydrogen Bonding of 1,2-Azaborines in the Binding Cavity of T4 Lysozyme Mutants: Structures and Thermodynamics.
J.Am.Chem.Soc., 138, 2016
|
|
5KHZ
 
 | | PSEUDO T4 LYSOZYME | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, Endolysin | | 著者 | Scholfield, M.R. | | 登録日 | 2016-06-16 | | 公開日 | 2017-04-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Structure-Energy Relationships of Halogen Bonds in Proteins.
Biochemistry, 56, 2017
|
|
5KI8
 
 | |
5JWW
 
 | | T4 Lysozyme L99A/M102Q with 1-Hydro-2-ethyl-1,2-azaborine Bound | | 分子名称: | 2-ethyl-1,2-dihydro-1,2-azaborinine, CHLORIDE ION, Endolysin | | 著者 | Lee, H, Fischer, M, Shoichet, B.K, Liu, S.-Y. | | 登録日 | 2016-05-12 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Hydrogen Bonding of 1,2-Azaborines in the Binding Cavity of T4 Lysozyme Mutants: Structures and Thermodynamics.
J.Am.Chem.Soc., 138, 2016
|
|
5KI1
 
 | | PSEUDO T4 LYSOZYME MUTANT - Y18F | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, Endolysin | | 著者 | Scholfield, M.R. | | 登録日 | 2016-06-16 | | 公開日 | 2017-04-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Structure-Energy Relationships of Halogen Bonds in Proteins.
Biochemistry, 56, 2017
|
|
5KI2
 
 | |
5KI3
 
 | |
5KGR
 
 | | Spin-Labeled T4 Lysozyme Construct I9V1/V131V1 (30 days) | | 分子名称: | CHLORIDE ION, Endolysin, HEXANE-1,6-DIOL, ... | | 著者 | Balo, A.R, Feyrer, H, Ernst, O.P. | | 登録日 | 2016-06-13 | | 公開日 | 2017-02-15 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.473 Å) | | 主引用文献 | Toward Precise Interpretation of DEER-Based Distance Distributions: Insights from Structural Characterization of V1 Spin-Labeled Side Chains.
Biochemistry, 55, 2016
|
|
5KIG
 
 | | PSEUDO T4 LYSOZYME MUTANT - Y88F | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, Endolysin | | 著者 | Scholfield, M.R. | | 登録日 | 2016-06-16 | | 公開日 | 2017-04-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure-Energy Relationships of Halogen Bonds in Proteins.
Biochemistry, 56, 2017
|
|
5KII
 
 | |
5KIM
 
 | | PSEUDO T4 LYSOZYME MUTANT - Y88PHE-I | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, Endolysin, SODIUM ION | | 著者 | Scholfield, M.R. | | 登録日 | 2016-06-16 | | 公開日 | 2017-04-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure-Energy Relationships of Halogen Bonds in Proteins.
Biochemistry, 56, 2017
|
|
2QB0
 
 | |
4W52
 
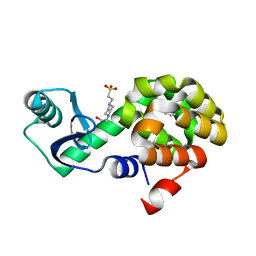 | | T4 Lysozyme L99A with Benzene Bound | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BENZENE, Endolysin | | 著者 | Merski, M, Shoichet, B.K, Eidam, O, Fischer, M. | | 登録日 | 2014-08-16 | | 公開日 | 2015-04-01 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.5001 Å) | | 主引用文献 | Homologous ligands accommodated by discrete conformations of a buried cavity.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4W54
 
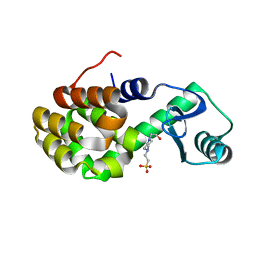 | | T4 Lysozyme L99A with Ethylbenzene Bound | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Endolysin, PHENYLETHANE | | 著者 | Merski, M, Shoichet, B.K, Eidam, O, Fischer, M. | | 登録日 | 2014-08-16 | | 公開日 | 2015-04-01 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.7901 Å) | | 主引用文献 | Homologous ligands accommodated by discrete conformations of a buried cavity.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4W57
 
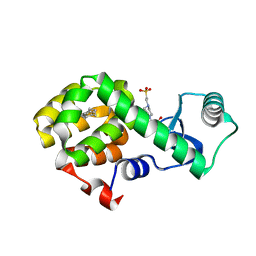 | | T4 Lysozyme L99A with n-Butylbenzene Bound | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Endolysin, N-BUTYLBENZENE | | 著者 | Merski, M, Shoichet, B.K, Eidam, O, Fischer, M. | | 登録日 | 2014-08-16 | | 公開日 | 2015-04-01 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.6801 Å) | | 主引用文献 | Homologous ligands accommodated by discrete conformations of a buried cavity.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
