1PAR
 
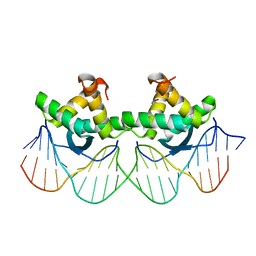 | | DNA RECOGNITION BY BETA-SHEETS IN THE ARC REPRESSOR-OPERATOR CRYSTAL STRUCTURE | | 分子名称: | DNA (5'-D(*AP*AP*TP*GP*AP*TP*AP*GP*AP*AP*GP*CP*AP*CP*TP*CP*T P*AP*CP*TP*AP*T)- 3'), DNA (5'-D(*TP*AP*TP*AP*GP*TP*AP*GP*AP*GP*TP*GP*CP*TP*TP*CP*T P*AP*TP*CP*AP*T)- 3'), PROTEIN (ARC REPRESSOR) | | 著者 | Raumann, B.E, Rould, M.A, Pabo, C.O, Sauer, R.T. | | 登録日 | 1994-03-22 | | 公開日 | 1994-07-31 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | DNA recognition by beta-sheets in the Arc repressor-operator crystal structure.
Nature, 367, 1994
|
|
1SV0
 
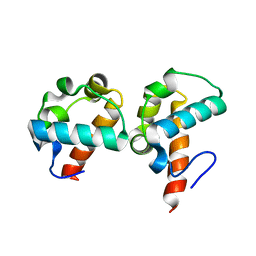 | | Crystal Structure Of Yan-SAM/Mae-SAM Complex | | 分子名称: | Ets DNA-binding protein pokkuri, modulator of the activity of Ets CG15085-PA | | 著者 | Qiao, F, Song, H, Kim, C.A, Sawaya, M.R, Hunter, J.B, Gingery, M, Rebay, I, Courey, A.J, Bowie, J.U. | | 登録日 | 2004-03-26 | | 公開日 | 2004-07-27 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Derepression by depolymerization; structural insights into the regulation of yan by mae.
Cell(Cambridge,Mass.), 118, 2004
|
|
1SV4
 
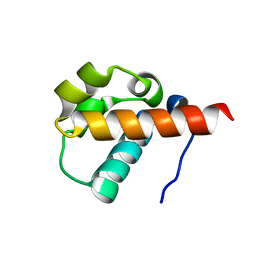 | | Crystal Structure of Yan-SAM | | 分子名称: | Ets DNA-binding protein pokkuri | | 著者 | Qiao, F, Song, H, Kim, C.A, Sawaya, M.R, Hunter, J.B, Gingery, M, Rebay, I, Courey, A.J, Bowie, J.U. | | 登録日 | 2004-03-27 | | 公開日 | 2004-07-27 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Derepression by depolymerization; structural insights into the regulation of yan by mae.
Cell(Cambridge,Mass.), 118, 2004
|
|
5GEP
 
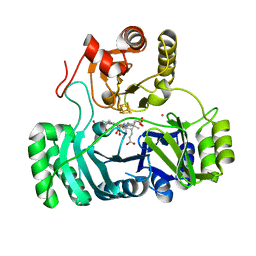 | | SULFITE REDUCTASE HEMOPROTEIN CARBON MONOXIDE COMPLEX REDUCED WITH CRII EDTA | | 分子名称: | CARBON MONOXIDE, IRON/SULFUR CLUSTER, POTASSIUM ION, ... | | 著者 | Crane, B.R, Getzoff, E.D. | | 登録日 | 1997-07-10 | | 公開日 | 1998-01-14 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Probing the catalytic mechanism of sulfite reductase by X-ray crystallography: structures of the Escherichia coli hemoprotein in complex with substrates, inhibitors, intermediates, and products.
Biochemistry, 36, 1997
|
|
3GEO
 
 | | SULFITE REDUCTASE HEMOPROTEIN NITRITE COMPLEX | | 分子名称: | IRON/SULFUR CLUSTER, NITRITE ION, SIROHEME, ... | | 著者 | Crane, B.R, Getzoff, E.D. | | 登録日 | 1997-07-10 | | 公開日 | 1998-01-14 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Probing the catalytic mechanism of sulfite reductase by X-ray crystallography: structures of the Escherichia coli hemoprotein in complex with substrates, inhibitors, intermediates, and products.
Biochemistry, 36, 1997
|
|
6GEP
 
 | | SULFITE REDUCTASE HEMOPROTEIN NITRIC OXIDE COMPLEX REDUCED WITH PROFLAVINE EDTA | | 分子名称: | IRON/SULFUR CLUSTER, NITRIC OXIDE, POTASSIUM ION, ... | | 著者 | Crane, B.R, Getzoff, E.D. | | 登録日 | 1997-07-10 | | 公開日 | 1998-01-14 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Probing the catalytic mechanism of sulfite reductase by X-ray crystallography: structures of the Escherichia coli hemoprotein in complex with substrates, inhibitors, intermediates, and products.
Biochemistry, 36, 1997
|
|
3AYJ
 
 | | X-ray crystal structures of L-phenylalanine oxidase (deaminating and decaboxylating) from Pseudomonas sp. P501. Structures of the enzyme-ligand complex and catalytic mechanism | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, PHENYLALANINE, ... | | 著者 | Ida, K, Suguro, M, Suzuki, H. | | 登録日 | 2011-05-07 | | 公開日 | 2011-08-31 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | High resolution X-ray crystal structures of L-phenylalanine oxidase (deaminating and decarboxylating) from Pseudomonas sp. P-501. Structures of the enzyme-ligand complex and catalytic mechanism
J.Biochem., 150, 2011
|
|
3AYL
 
 | |
3AYI
 
 | | X-ray crystal structures of L-phenylalanine oxidase (deaminating and decaboxylating) from Pseudomonas sp. P501. Structures of the enzyme-ligand complex and catalytic mechanism | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, HYDROCINNAMIC ACID, ... | | 著者 | Ida, K, Suguro, M, Suzuki, H. | | 登録日 | 2011-05-07 | | 公開日 | 2011-08-31 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | High resolution X-ray crystal structures of L-phenylalanine oxidase (deaminating and decarboxylating) from Pseudomonas sp. P-501. Structures of the enzyme-ligand complex and catalytic mechanism
J.Biochem., 150, 2011
|
|
8B0U
 
 | |
2GEP
 
 | | SULFITE REDUCTASE HEMOPROTEIN, OXIDIZED, SIROHEME FEIII [4FE-4S] +2,SULFITE COMPLEX | | 分子名称: | IRON/SULFUR CLUSTER, SIROHEME, SODIUM ION, ... | | 著者 | Crane, B.R, Getzoff, E.D. | | 登録日 | 1997-07-10 | | 公開日 | 1998-01-14 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Probing the catalytic mechanism of sulfite reductase by X-ray crystallography: structures of the Escherichia coli hemoprotein in complex with substrates, inhibitors, intermediates, and products.
Biochemistry, 36, 1997
|
|
2YR5
 
 | | Crystal structure of L-phenylalanine oxidase from Psuedomonas sp.P501 | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Pro-enzyme of L-phenylalanine oxidase, ... | | 著者 | Ida, K, Kurabayashi, M, Suguro, M, Hikima, T, Yamamoto, M, Suzuki, H. | | 登録日 | 2007-04-02 | | 公開日 | 2008-04-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Structural basis of proteolytic activation of L-phenylalanine oxidase from Pseudomonas sp. P-501.
J.Biol.Chem., 283, 2008
|
|
8GEP
 
 | | SULFITE REDUCTASE HEMOPROTEIN NITRATE COMPLEX | | 分子名称: | IRON/SULFUR CLUSTER, NITRATE ION, POTASSIUM ION, ... | | 著者 | Crane, B.R, Getzoff, E.D. | | 登録日 | 1997-07-11 | | 公開日 | 1998-01-14 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Probing the catalytic mechanism of sulfite reductase by X-ray crystallography: structures of the Escherichia coli hemoprotein in complex with substrates, inhibitors, intermediates, and products.
Biochemistry, 36, 1997
|
|
2YR4
 
 | | Crystal structure of L-phenylalanine oxiase from Psuedomonas sp. P-501 | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Pro-enzyme of L-phenylalanine oxidase, SULFATE ION | | 著者 | Ida, K, Kurabayashi, M, Suguro, M, Suzuki, H. | | 登録日 | 2007-04-02 | | 公開日 | 2008-04-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural basis of proteolytic activation of L-phenylalanine oxidase from Pseudomonas sp. P-501.
J.Biol.Chem., 283, 2008
|
|
2YR6
 
 | | Crystal structure of L-phenylalanine oxidase from Psuedomonas sp.P501 | | 分子名称: | 2-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | 著者 | Ida, K, Kurabayashi, M, Suguro, M, Suzuki, H. | | 登録日 | 2007-04-02 | | 公開日 | 2008-04-15 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Structural basis of proteolytic activation of L-phenylalanine oxidase from Pseudomonas sp. P-501.
J.Biol.Chem., 283, 2008
|
|
4GEP
 
 | | SULFITE REDUCTASE HEMOPROTEIN CYANIDE COMPLEX REDUCED WITH CRII EDTA | | 分子名称: | CYANIDE ION, IRON/SULFUR CLUSTER, POTASSIUM ION, ... | | 著者 | Crane, B.R, Getzoff, E.D. | | 登録日 | 1997-07-10 | | 公開日 | 1998-01-14 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Probing the catalytic mechanism of sulfite reductase by X-ray crystallography: structures of the Escherichia coli hemoprotein in complex with substrates, inhibitors, intermediates, and products.
Biochemistry, 36, 1997
|
|
7GEP
 
 | | SULFITE REDUCTASE HEMOPROTEIN IN COMPLEX WITH A PARTIALLY OXIDIZED SULFIDE SPECIES | | 分子名称: | IRON/SULFUR CLUSTER, SIROHEME, SODIUM ION, ... | | 著者 | Crane, B.R, Getzoff, E.D. | | 登録日 | 1997-07-11 | | 公開日 | 1998-01-14 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Probing the catalytic mechanism of sulfite reductase by X-ray crystallography: structures of the Escherichia coli hemoprotein in complex with substrates, inhibitors, intermediates, and products.
Biochemistry, 36, 1997
|
|
6TVE
 
 | |
6TVG
 
 | |
8B0R
 
 | | Structure of the CalpL/cA4 complex | | 分子名称: | Cyclic tetraadenosine monophosphate (cA4), SMODS-associated and fused to various effectors domain-containing protein, SULFATE ION, ... | | 著者 | Schneberger, N, Hagelueken, G. | | 登録日 | 2022-09-08 | | 公開日 | 2022-11-16 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Antiviral signalling by a cyclic nucleotide activated CRISPR protease.
Nature, 614, 2023
|
|
1ZJ9
 
 | | Structure of Mycobacterium tuberculosis NirA protein | | 分子名称: | CHLORIDE ION, IRON/SULFUR CLUSTER, Probable ferredoxin-dependent nitrite reductase NirA, ... | | 著者 | Schnell, R, Sandalova, T, Hellman, U, Lindqvist, Y, Schneider, G. | | 登録日 | 2005-04-28 | | 公開日 | 2005-05-31 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Siroheme- and [Fe4-S4]-dependent NirA from Mycobacterium tuberculosis Is a Sulfite Reductase with a Covalent Cys-Tyr Bond in the Active Site
J.Biol.Chem., 280, 2005
|
|
1ZJ8
 
 | | Structure of Mycobacterium tuberculosis NirA protein | | 分子名称: | CHLORIDE ION, IRON/SULFUR CLUSTER, Probable ferredoxin-dependent nitrite reductase NirA, ... | | 著者 | Schnell, R, Sandalova, T, Hellman, U, Lindqvist, Y, Schneider, G, Structural Proteomics in Europe (SPINE) | | 登録日 | 2005-04-28 | | 公開日 | 2005-05-31 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Siroheme- and [Fe4-S4]-dependent NirA from Mycobacterium tuberculosis Is a Sulfite Reductase with a Covalent Cys-Tyr Bond in the Active Site
J.Biol.Chem., 280, 2005
|
|
7X13
 
 | |
5FGP
 
 | | Crystal structure of D. melanogaster Pur-alpha repeat I-II in complex with DNA. | | 分子名称: | CG1507-PB, isoform B, CHLORIDE ION, ... | | 著者 | Weber, J, Janowski, R, Niessing, D. | | 登録日 | 2015-12-21 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis of nucleic-acid recognition and double-strand unwinding by the essential neuronal protein Pur-alpha.
Elife, 5, 2016
|
|
5FGO
 
 | |
