5RPK
 
 | | PanDDA analysis group deposition -- Proteinase K changed state model for fragment Frag Xtal Screen B9a | | 分子名称: | Proteinase K, SULFATE ION, piperidine-1-carboximidamide | | 著者 | Lima, G.M.A, Talibov, V, Benz, L.S, Jagudin, E, Mueller, U. | | 登録日 | 2020-09-23 | | 公開日 | 2021-05-26 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | FragMAXapp: crystallographic fragment-screening data-analysis and project-management system.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5RPM
 
 | | PanDDA analysis group deposition -- Proteinase K changed state model for fragment Frag Xtal Screen H5a | | 分子名称: | 4-HYDROXYBENZAMIDE, Proteinase K, SULFATE ION | | 著者 | Lima, G.M.A, Talibov, V, Benz, L.S, Jagudin, E, Mueller, U. | | 登録日 | 2020-09-23 | | 公開日 | 2021-05-26 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.22 Å) | | 主引用文献 | FragMAXapp: crystallographic fragment-screening data-analysis and project-management system.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5RPN
 
 | | PanDDA analysis group deposition -- Proteinase K crystal structure Apo64 | | 分子名称: | Proteinase K, SULFATE ION | | 著者 | Lima, G.M.A, Talibov, V, Benz, L.S, Jagudin, E, Mueller, U. | | 登録日 | 2020-09-23 | | 公開日 | 2021-05-26 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.02 Å) | | 主引用文献 | FragMAXapp: crystallographic fragment-screening data-analysis and project-management system.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5RPO
 
 | | PanDDA analysis group deposition -- Proteinase K crystal structure Apo7 | | 分子名称: | Proteinase K, SULFATE ION | | 著者 | Lima, G.M.A, Talibov, V, Benz, L.S, Jagudin, E, Mueller, U. | | 登録日 | 2020-09-23 | | 公開日 | 2021-05-26 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | FragMAXapp: crystallographic fragment-screening data-analysis and project-management system.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5RPW
 
 | | PanDDA analysis group deposition -- Proteinase K crystal structure Apo63 | | 分子名称: | Proteinase K | | 著者 | Lima, G.M.A, Talibov, V, Benz, L.S, Jagudin, E, Mueller, U. | | 登録日 | 2020-09-23 | | 公開日 | 2021-05-26 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.02 Å) | | 主引用文献 | FragMAXapp: crystallographic fragment-screening data-analysis and project-management system.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5ROR
 
 | | PanDDA analysis group deposition -- Proteinase K changed state model for fragment Frag Xtal Screen F1a | | 分子名称: | NICOTINAMIDE, Proteinase K, SULFATE ION | | 著者 | Lima, G.M.A, Talibov, V, Benz, L.S, Jagudin, E, Mueller, U. | | 登録日 | 2020-09-23 | | 公開日 | 2021-05-26 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.22 Å) | | 主引用文献 | FragMAXapp: crystallographic fragment-screening data-analysis and project-management system.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5ROS
 
 | | PanDDA analysis group deposition -- Proteinase K crystal structure Apo34 | | 分子名称: | Proteinase K, SULFATE ION | | 著者 | Lima, G.M.A, Talibov, V, Benz, L.S, Jagudin, E, Mueller, U. | | 登録日 | 2020-09-23 | | 公開日 | 2021-05-26 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | FragMAXapp: crystallographic fragment-screening data-analysis and project-management system.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5RP4
 
 | | PanDDA analysis group deposition -- Proteinase K crystal structure Apo70 | | 分子名称: | Proteinase K, SULFATE ION | | 著者 | Lima, G.M.A, Talibov, V, Benz, L.S, Jagudin, E, Mueller, U. | | 登録日 | 2020-09-23 | | 公開日 | 2021-05-26 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | FragMAXapp: crystallographic fragment-screening data-analysis and project-management system.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5RPR
 
 | | PanDDA analysis group deposition -- Proteinase K crystal structure Apo15 | | 分子名称: | Proteinase K, SULFATE ION | | 著者 | Lima, G.M.A, Talibov, V, Benz, L.S, Jagudin, E, Mueller, U. | | 登録日 | 2020-09-23 | | 公開日 | 2021-05-26 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.16 Å) | | 主引用文献 | FragMAXapp: crystallographic fragment-screening data-analysis and project-management system.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5RPG
 
 | | PanDDA analysis group deposition -- Proteinase K changed state model for fragment Frag Xtal Screen H2a | | 分子名称: | Proteinase K, SULFATE ION, TRANS-4-AMINOMETHYLCYCLOHEXANE-1-CARBOXYLIC ACID | | 著者 | Lima, G.M.A, Talibov, V, Benz, L.S, Jagudin, E, Mueller, U. | | 登録日 | 2020-09-23 | | 公開日 | 2021-05-26 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | FragMAXapp: crystallographic fragment-screening data-analysis and project-management system.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5XGO
 
 | | The Ferritin E-Domain: Toward Understanding Its Role in Protein Cage Assembly Through the Crystal Structure of a Maxi-/Mini-Ferritin Chimera | | 分子名称: | CHLORIDE ION, DNA protection during starvation protein,Bacterioferritin | | 著者 | Cornell, T.A, Srivastava, Y, Jauch, R, Fan, R, Orner, B.P. | | 登録日 | 2017-04-14 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | The Crystal Structure of a Maxi/Mini-Ferritin Chimera Reveals Guiding Principles for the Assembly of Protein Cages.
Biochemistry, 56, 2017
|
|
6M1H
 
 | | CryoEM structure of human PAC1 receptor in complex with maxadilan | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | 著者 | Song, X, Wang, J, Zhang, D, Wang, H.W, Ma, Y. | | 登録日 | 2020-02-26 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM structures of PAC1 receptor reveal ligand binding mechanism.
Cell Res., 30, 2020
|
|
1F1C
 
 | | CRYSTAL STRUCTURE OF CYTOCHROME C549 | | 分子名称: | CYTOCHROME C549, HEME C | | 著者 | Kerfeld, C.A, Sawaya, M.R, Yeates, T.O, Krogmann, D.W. | | 登録日 | 2000-05-18 | | 公開日 | 2001-08-08 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structures of cytochrome c-549 and cytochrome c6 from the cyanobacterium Arthrospira maxima.
Biochemistry, 40, 2001
|
|
3C7A
 
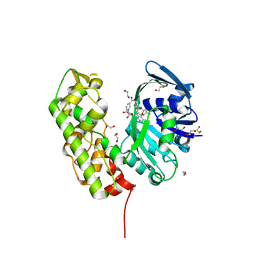 | | A structural basis for substrate and stereo selectivity in octopine dehydrogenase (ODH-NADH) | | 分子名称: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Octopine dehydrogenase | | 著者 | Smits, S.H.J, Mueller, A, Schmitt, L, Grieshaber, M.K. | | 登録日 | 2008-02-07 | | 公開日 | 2008-07-22 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A structural basis for substrate selectivity and stereoselectivity in octopine dehydrogenase from Pecten maximus.
J.Mol.Biol., 381, 2008
|
|
3C7D
 
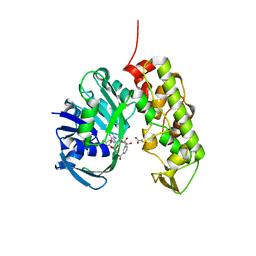 | | A structural basis for substrate and stereo selectivity in octopine dehydrogenase (ODH-NADH-Pyruvate) | | 分子名称: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Octopine dehydrogenase, PYRUVIC ACID | | 著者 | Smits, S.H.J, Mueller, A, Schmitt, L, Grieshaber, M.K. | | 登録日 | 2008-02-07 | | 公開日 | 2008-07-22 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A structural basis for substrate selectivity and stereoselectivity in octopine dehydrogenase from Pecten maximus.
J.Mol.Biol., 381, 2008
|
|
3C7C
 
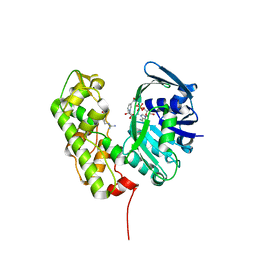 | | A structural basis for substrate and stereo selectivity in octopine dehydrogenase (ODH-NADH-L-Arginine) | | 分子名称: | ARGININE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Octopine dehydrogenase | | 著者 | Smits, S.H.J, Mueller, A, Schmitt, L, Grieshaber, M.K. | | 登録日 | 2008-02-07 | | 公開日 | 2008-07-22 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | A structural basis for substrate selectivity and stereoselectivity in octopine dehydrogenase from Pecten maximus.
J.Mol.Biol., 381, 2008
|
|
3B0T
 
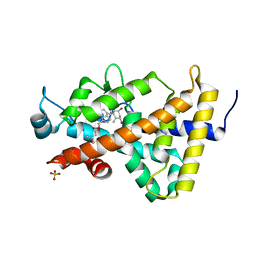 | | Human VDR ligand binding domain in complex with maxacalcitol | | 分子名称: | (1S,3R,5Z,7E,14beta,17alpha,20S)-20-(3-hydroxy-3-methylbutoxy)-9,10-secopregna-5,7,10-triene-1,3-diol, SULFATE ION, Vitamin D3 receptor | | 著者 | Hishiki, A, Hashimoto, H, Sato, M, Shimizu, T. | | 登録日 | 2011-06-14 | | 公開日 | 2011-08-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Human VDR ligand binding domain in complex with maxacalcitol
To be Published
|
|
1EMY
 
 | |
3IQD
 
 | | Structure of Octopine-dehydrogenase in complex with NADH and Agmatine | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, AGMATINE, Octopine dehydrogenase | | 著者 | Smits, S.H.J, Meyer, T, Mueller, A, Willbold, D, Grieshaber, M.K, Schmitt, L. | | 登録日 | 2009-08-20 | | 公開日 | 2010-08-25 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Insights into the mechanism of ligand binding to octopine dehydrogenase from Pecten maximus by NMR and crystallography
Plos One, 5, 2010
|
|
2N1E
 
 | |
1PPE
 
 | |
1SMR
 
 | |
1PC6
 
 | | Structural Genomics, NinB | | 分子名称: | BETA-MERCAPTOETHANOL, Protein ninB | | 著者 | Zhang, R, Beasley, S, Maxwell, K.L, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2003-05-15 | | 公開日 | 2004-01-20 | | 最終更新日 | 2025-03-26 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Functional similarities between phage lambda Orf and Escherichia coli RecFOR in initiation of genetic exchange
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
4PMS
 
 | | The structure of TrkA kinase bound to the inhibitor 4-naphthalen-1-yl-1-[(5-phenyl-1,2,4-oxadiazol-3-yl)methyl]-1H-pyrrolo[3,2-c]pyridine-2-carboxylic acid | | 分子名称: | 4-(naphthalen-1-yl)-1-[(5-phenyl-1,2,4-oxadiazol-3-yl)methyl]-1H-pyrrolo[3,2-c]pyridine-2-carboxylic acid, ACETATE ION, CHLORIDE ION, ... | | 著者 | Su, H.P. | | 登録日 | 2014-05-22 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Maximizing diversity from a kinase screen: identification of novel and selective pan-Trk inhibitors for chronic pain.
J.Med.Chem., 57, 2014
|
|
4PMM
 
 | |
