1GVX
 
 | | Endothiapepsin complexed with H256 | | 分子名称: | ENDOTHIAPEPSIN, INHIBITOR H256, SULFATE ION | | 著者 | Coates, L, Erskine, P.T, Crump, M.P, Wood, S.P, Cooper, J.B. | | 登録日 | 2002-02-27 | | 公開日 | 2002-07-04 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Five Atomic Resolution Structures of Endothiapepsin Inhibitor Complexes: Implications for the Aspartic Proteinase Mechanism
J.Mol.Biol., 318, 2002
|
|
1GVW
 
 | | Endothiapepsin complex with PD-130,328 | | 分子名称: | ENDOTHIAPEPSIN, N-(tert-butoxycarbonyl)-L-phenylalanyl-N-{(1S)-1-[(R)-hydroxy(2-{[(2S)-2-methylbutyl]amino}-2-oxoethyl)phosphoryl]-3-methylbutyl}-3-(1H-imidazol-3-ium-4-yl)-L-alaninamide, SULFATE ION | | 著者 | Coates, L, Erskine, P.T, Crump, M.P, Wood, S.P, Cooper, J.B. | | 登録日 | 2002-02-27 | | 公開日 | 2002-07-04 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Five Atomic Resolution Structures of Endothiapepsin Inhibitor Complexes: Implications for the Aspartic Proteinase Mechanism
J.Mol.Biol., 318, 2002
|
|
1HH3
 
 | | Decaplanin first P21-Form | | 分子名称: | 4-epi-vancosamine, DECAPLANIN, GLYCEROL, ... | | 著者 | Lehmann, C, Vertessy, L, Sheldrick, G.M, Dauter, Z, Dauter, M. | | 登録日 | 2000-12-19 | | 公開日 | 2005-07-11 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Structures of Four Crystal Forms of Decaplanin
Helv.Chim.Acta, 86, 2003
|
|
2X46
 
 | | Crystal Structure of SeMet Arg r 1 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ALLERGEN ARG R 1 | | 著者 | Paesen, G.C, Siebold, C, Syme, N, Harlos, K, Graham, S.C, Hilger, C, Homans, S.W, Hentges, F, Stuart, D.I. | | 登録日 | 2010-01-28 | | 公開日 | 2011-02-09 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Crystal Structure of the Allergen Arg R 1, a Histamine-Binding Lipocalin from a Soft Tick
To be Published
|
|
5RT7
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000015442276 | | 分子名称: | 1H-PYRROLO[2,3-B]PYRIDINE, Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-09-28 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RTO
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000388302 | | 分子名称: | 4-PIPERIDINO-PIPERIDINE, Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-09-28 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RU6
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000001442764 | | 分子名称: | Non-structural protein 3, naphthalene-2-carboximidamide | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-09-28 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RUO
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000001683100 | | 分子名称: | 4-chloro-1H-indole-2-carboxylic acid, Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-09-28 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
2R31
 
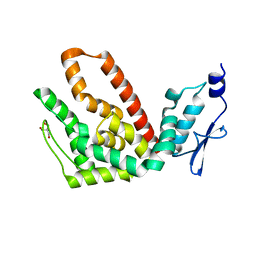 | | Crystal structure of atp12p from paracoccus denitrificans | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ATP12 ATPase | | 著者 | Ludlam, A.V, Brunzelle, J.S, Gatti, D.L, Ackerman, S.H. | | 登録日 | 2007-08-28 | | 公開日 | 2008-03-25 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Chaperones of F1-ATPase.
J.Biol.Chem., 284, 2009
|
|
6ODG
 
 | | SVQIVY, Crystal Structure of a tau protein fragment | | 分子名称: | Microtubule-associated protein tau | | 著者 | Eisenberg, D.S, Boyer, D.R, Sawaya, M.R, Seidler, P.M. | | 登録日 | 2019-03-26 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Structure-based inhibitors halt prion-like seeding by Alzheimer's disease-and tauopathy-derived brain tissue samples.
J.Biol.Chem., 294, 2019
|
|
1QYL
 
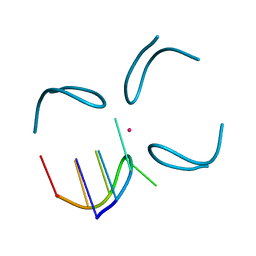 | | GCATGCT + Vanadium | | 分子名称: | 5'-D(*GP*CP*AP*TP*GP*CP*T)-3', VANADIUM ION | | 著者 | Cardin, C.J, Gan, Y, Thorpe, J.H, Teixeira, S.C.M, Gale, B.C, Moraes, M.I.A. | | 登録日 | 2003-09-11 | | 公開日 | 2003-10-21 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Metal Ion Distribution and Stabilisation of the DNA Quadruplex Structure Formed by d(GCATGCT)
To be Published
|
|
2PZN
 
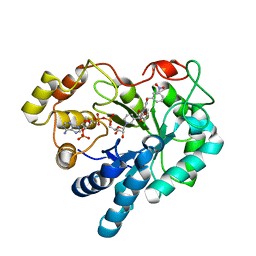 | | The crystallographic structure of Aldose Reductase IDD393 complex confirms Leu300 as a specificity determinant | | 分子名称: | (5-CHLORO-2-{[(3-NITROBENZYL)AMINO]CARBONYL}PHENOXY)ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Ruiz, F, Hazemann, I, Darmanin, C, Mitschler, A, Van Zandt, M, Joachimiak, A, El-Kabbani, O, Podjarny, A. | | 登録日 | 2007-05-18 | | 公開日 | 2008-05-27 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | The Crystallographic Structure of Alr2-Idd393 Complex Confirms Leu300 as a Specificity Determinant
To be Published
|
|
6X7T
 
 | |
6X9M
 
 | |
6X7Z
 
 | | Inositol-bound structure of Marinomonas primoryensis PA14 carbohydrate-binding domain | | 分子名称: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1,2-ETHANEDIOL, Antifreeze protein, ... | | 著者 | Guo, S, Davies, P.L. | | 登録日 | 2020-06-01 | | 公開日 | 2021-06-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Structural Basis of Ligand Selectivity by a Bacterial Adhesin Lectin Involved in Multispecies Biofilm Formation.
Mbio, 12, 2021
|
|
6X9P
 
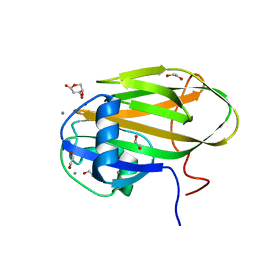 | |
6X7Y
 
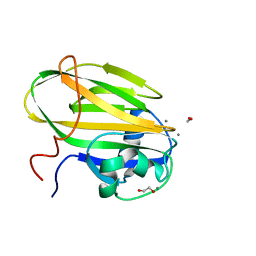 | |
2CNQ
 
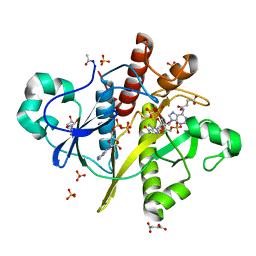 | | Atomic resolution structure of SAICAR-synthase from Saccharomyces cerevisiae complexed with ADP, AICAR, succinate | | 分子名称: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, ... | | 著者 | Urusova, D.V, Antonyuk, S.V, Grebenko, A.I, Levdikov, V.M, Barynin, V.V, Popov, A.N, Lamzin, V.S, Melik-Adamyan, W.R. | | 登録日 | 2006-05-23 | | 公開日 | 2006-06-08 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Saicar Synthase: Substrate Recognition, Conformational Flexibility and Catalysis.
To be Published
|
|
8C10
 
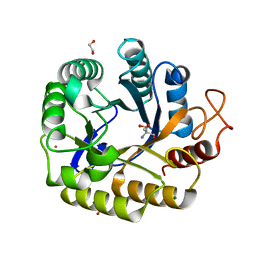 | | Biochemical and structural characterisation of an alkaline family GH5 cellulase from a shipworm symbiont | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GH5 Cellulase, ... | | 著者 | Leiros, I, Vaaje-Kolstad, G. | | 登録日 | 2022-12-19 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Biochemical and structural characterisation of a family GH5 cellulase from endosymbiont of shipworm P. megotara.
Biotechnol Biofuels Bioprod, 16, 2023
|
|
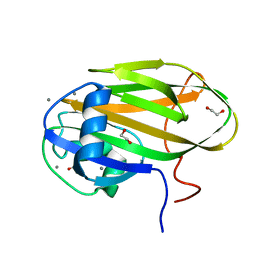
6X8Y
 
 | |
6X8D
 
 | |
5HQ1
 
 | | Comment on S. W. M. Tanley and J. R. Helliwell Structural dynamics of cisplatin binding to histidine in a protein Struct. Dyn. 1, 034701 (2014) regarding the refinement of 4mwk, 4mwm, 4mwn and 4oxe and the method we have adopted. | | 分子名称: | CHLORIDE ION, DIMETHYL SULFOXIDE, Lysozyme C, ... | | 著者 | Helliwell, J.R. | | 登録日 | 2016-01-21 | | 公開日 | 2016-05-18 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Comment on "Structural dynamics of cisplatin binding to histidine in a protein" [Struct. Dyn. 1, 034701 (2014)].
Struct Dyn, 3, 2016
|
|
4DPB
 
 | | The 1.00 Angstrom crystal structure of oxidized (CuII) poplar plastocyanin A at pH 8.0 | | 分子名称: | COPPER (II) ION, Plastocyanin A, chloroplastic | | 著者 | Kachalova, G.S, Shosheva, A.H, Bourenkov, G.P, Donchev, A.A, Dimitrov, M.I, Bartunik, H.D. | | 登録日 | 2012-02-13 | | 公開日 | 2013-02-13 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Structural comparison of the poplar plastocyanin isoforms PCa and PCb sheds new light on the role of the copper site geometry in interactions with redox partners in oxygenic photosynthesis.
J.Inorg.Biochem., 115, 2012
|
|
6ZX1
 
 | | OMPD-domain of human UMPS in complex with 6-Aza-UMP at 1.0 Angstroms resolution | | 分子名称: | 6-AZA URIDINE 5'-MONOPHOSPHATE, PROLINE, SULFATE ION, ... | | 著者 | Tittmann, K, Rindfleisch, S, Krull, M. | | 登録日 | 2020-07-29 | | 公開日 | 2022-02-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6ZWY
 
 | |
