5JWW
 
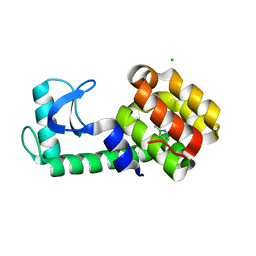 | | T4 Lysozyme L99A/M102Q with 1-Hydro-2-ethyl-1,2-azaborine Bound | | 分子名称: | 2-ethyl-1,2-dihydro-1,2-azaborinine, CHLORIDE ION, Endolysin | | 著者 | Lee, H, Fischer, M, Shoichet, B.K, Liu, S.-Y. | | 登録日 | 2016-05-12 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Hydrogen Bonding of 1,2-Azaborines in the Binding Cavity of T4 Lysozyme Mutants: Structures and Thermodynamics.
J.Am.Chem.Soc., 138, 2016
|
|
5JY7
 
 | |
7XZZ
 
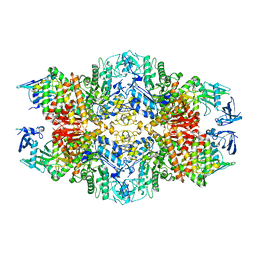
 | | Cryo-EM structure of the nucleosome in complex with p53 | | 分子名称: | Cellular tumor antigen p53, DNA (169-MER), Histone H2A type 1-B/E, ... | | 著者 | Nishimura, M, Nozawa, K, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2022-06-03 | | 公開日 | 2022-10-12 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (4.07 Å) | | 主引用文献 | Structural basis for p53 binding to its nucleosomal target DNA sequence.
Pnas Nexus, 1, 2022
|
|
5KAF
 
 | | RT XFEL structure of Photosystem II in the dark state at 3.0 A resolution | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | 登録日 | 2016-06-01 | | 公開日 | 2016-11-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.00001 Å) | | 主引用文献 | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
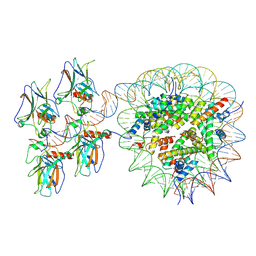
5K2P
 
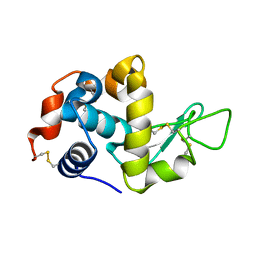 | |
7XWH
 
 | | structure of patulin-detoxifying enzyme with NADP+ | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | 著者 | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | 登録日 | 2022-05-26 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
7XWJ
 
 | | structure of patulin-detoxifying enzyme Y155F with NADPH | | 分子名称: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | 著者 | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | 登録日 | 2022-05-26 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
7XWL
 
 | | structure of patulin-detoxifying enzyme Y155F/V187F with NADPH | | 分子名称: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | 著者 | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | 登録日 | 2022-05-26 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
7XWI
 
 | | structure of patulin-detoxifying enzyme with NADPH | | 分子名称: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | 著者 | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | 登録日 | 2022-05-26 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
5KAR
 
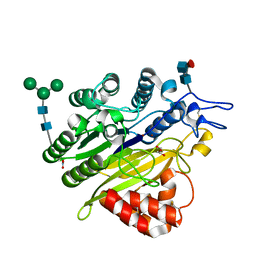 | | Murine acid sphingomyelinase-like phosphodiesterase 3b (SMPDL3B) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acid sphingomyelinase-like phosphodiesterase 3b, ... | | 著者 | Gorelik, A, Illes, K, Heinz, L.X, Superti-Furga, G, Nagar, B. | | 登録日 | 2016-06-02 | | 公開日 | 2016-10-05 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.142 Å) | | 主引用文献 | Crystal Structure of the Acid Sphingomyelinase-like Phosphodiesterase SMPDL3B Provides Insights into Determinants of Substrate Specificity.
J.Biol.Chem., 291, 2016
|
|
5KCR
 
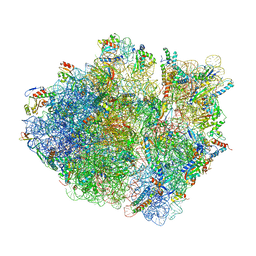 | | Cryo-EM structure of the Escherichia coli 70S ribosome in complex with antibiotic Avilamycin C, mRNA and P-site tRNA at 3.6A resolution | | 分子名称: | (2R,3S,4R,6S)-4-hydroxy-6-{[(2R,3aR,4R,4'R,5'S,6S,6'R,7aR)-4'-hydroxy-6-{[(2S,3R,4R,5S,6R)-3-hydroxy-2-{[(2R,3S,4S,5S,6S)-4-hydroxy-6-({(2R,3aS,3a'R,6S,6'R,7R,7'R,7aR,7a'R)-7'-hydroxy-7'-[(1S)-1-hydroxyethyl]-6'-methyl-7-[(2-methylpropanoyl)oxy]octahydro-4H-2,4'-spirobi[[1,3]dioxolo[4,5-c]pyran]-6-yl}oxy)-5-methoxy-2-(methoxymethyl)tetrahydro-2H-pyran-3-yl]oxy}-5-methoxy-6-methyltetrahydro-2H-pyran-4-yl]oxy}-4,6',7a-trimethyloctahydro-4H-spiro[1,3-dioxolo[4,5-c]pyran-2,2'-pyran]-5'-yl]oxy}-2-methyltetrahydro-2H-pyran-3-yl 3,5-dichloro-4-hydroxy-2-methoxy-6-methylbenzoate (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | 著者 | Arenz, S, Juette, M.F, Graf, M, Nguyen, F, Huter, P, Polikanov, Y.S, Blanchard, S.C, Wilson, D.N. | | 登録日 | 2016-06-06 | | 公開日 | 2016-08-17 | | 最終更新日 | 2019-12-25 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structures of the orthosomycin antibiotics avilamycin and evernimicin in complex with the bacterial 70S ribosome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7XWN
 
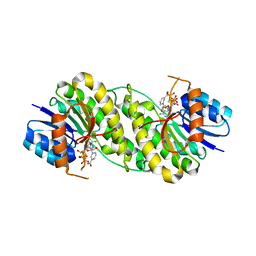 | | structure of patulin-detoxifying enzyme Y155F/V187K with NADPH and substrate | | 分子名称: | (4~{S})-4-oxidanyl-4,6-dihydrofuro[3,2-c]pyran-2-one, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | 著者 | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | 登録日 | 2022-05-26 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
7XWK
 
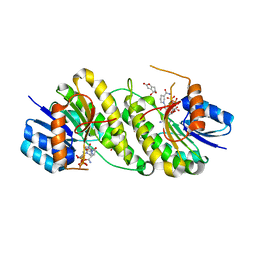 | | structure of patulin-detoxifying enzyme Y155F with NADPH and substrate | | 分子名称: | (4~{S})-4-oxidanyl-4,6-dihydrofuro[3,2-c]pyran-2-one, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | 著者 | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | 登録日 | 2022-05-26 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
5K9T
 
 | |
5KAG
 
 | | Crystal structure of a dioxygenase in the Crotonase superfamily in P21 | | 分子名称: | (3,5-dihydroxyphenyl)acetyl-CoA 1,2-dioxygenase, OXYGEN MOLECULE, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL (3R)-4-({3-[(2-{[(3,5-DIHYDROXYPHENYL)ACETYL]AMINO}ETHYL)AMINO]-3-OXOPROPYL}AMINO)-3-HYDROXY-2,2-DIMETHYL-4-OXOBUTYL DIHYDROGEN DIPHOSPHATE | | 著者 | Li, K, Fielding, E.N, Condurso, H.L, Bruner, S.D. | | 登録日 | 2016-06-01 | | 公開日 | 2017-06-21 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.456 Å) | | 主引用文献 | Probing the structural basis of oxygen binding in a cofactor-independent dioxygenase.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5KBP
 
 | | The crystal structure of an alpha-mannosidase from Enterococcus faecalis V583 | | 分子名称: | Glycosyl hydrolase, family 38, SULFATE ION | | 著者 | Tan, K, Chhor, G, Jedrzejczak, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-06-03 | | 公開日 | 2016-07-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The crystal structure of an alpha-mannosidase from Enterococcus faecalis V583
To Be Published
|
|
7YBO
 
 | | Crystal structure of FGFR4 kinase domain with 10z | | 分子名称: | Fibroblast growth factor receptor 4, SULFATE ION, ~{N}-[4-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-5-cyano-pyridin-2-yl]-6-bromanyl-5-(hydroxymethyl)-1-(2-morpholin-4-ylethyl)pyrrolo[3,2-b]pyridine-3-carboxamide | | 著者 | Chen, X.J, Lin, Q.M, Chen, Y.H. | | 登録日 | 2022-06-29 | | 公開日 | 2022-11-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.307 Å) | | 主引用文献 | Design, Synthesis, and Biological Evaluation of 5-Formyl-pyrrolo[3,2- b ]pyridine-3-carboxamides as New Selective, Potent, and Reversible-Covalent FGFR4 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7YC3
 
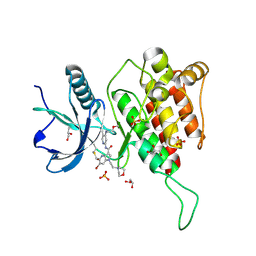 | | Crystal structure of FGFR4 kinase domain with 10t | | 分子名称: | 6-bromanyl-~{N}-[5-cyano-4-(2-methoxyethylamino)pyridin-2-yl]-5-methanoyl-1-(2-morpholin-4-ylethyl)pyrrolo[3,2-b]pyridine-3-carboxamide, Fibroblast growth factor receptor 4, GLYCEROL, ... | | 著者 | Chen, X.J, Lin, Q.M, Chen, Y.H. | | 登録日 | 2022-06-30 | | 公開日 | 2022-11-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.987 Å) | | 主引用文献 | Design, Synthesis, and Biological Evaluation of 5-Formyl-pyrrolo[3,2- b ]pyridine-3-carboxamides as New Selective, Potent, and Reversible-Covalent FGFR4 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7XUE
 
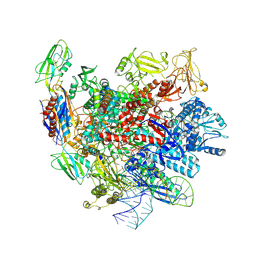 | |
7YC1
 
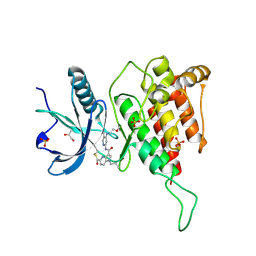 | | Crystal structure of FGFR4 kinase domain with 10d | | 分子名称: | Fibroblast growth factor receptor 4, GLYCEROL, SULFATE ION, ... | | 著者 | Chen, X.J, Lin, Q.M, Chen, Y.H. | | 登録日 | 2022-06-30 | | 公開日 | 2022-11-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.535 Å) | | 主引用文献 | Design, Synthesis, and Biological Evaluation of 5-Formyl-pyrrolo[3,2- b ]pyridine-3-carboxamides as New Selective, Potent, and Reversible-Covalent FGFR4 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7XUG
 
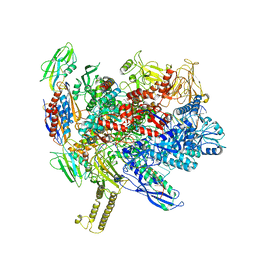 | |
7XUI
 
 | |
5KHD
 
 | | Structure of 1.75 A BldD C-domain-c-di-GMP complex | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DNA-binding protein | | 著者 | Schumacher, M. | | 登録日 | 2016-06-14 | | 公開日 | 2016-06-29 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.7501 Å) | | 主引用文献 | Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development.
Cell, 158, 2014
|
|
5KID
 
 | | Tightening the Recognition of Tetravalent Zr and Th Complexes by the Siderophore-Binding Mammalian Protein Siderocalin for Theranostic Applications | | 分子名称: | CHLORIDE ION, GLYCEROL, N,N'-(butane-1,4-diyl)bis(N-{3-[(2,3-dihydroxybenzene-1-carbonyl)amino]propyl}-2,3-dihydroxybenzamide), ... | | 著者 | Rupert, P.B, Strong, R.K. | | 登録日 | 2016-06-16 | | 公開日 | 2017-04-26 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Engineered Recognition of Tetravalent Zirconium and Thorium by Chelator-Protein Systems: Toward Flexible Radiotherapy and Imaging Platforms.
Inorg Chem, 55, 2016
|
|
7YBP
 
 | | Crystal structure of FGFR4(V550L) kinase domain with 10z | | 分子名称: | Fibroblast growth factor receptor 4, SULFATE ION, ~{N}-[4-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-5-cyano-pyridin-2-yl]-6-bromanyl-5-(hydroxymethyl)-1-(2-morpholin-4-ylethyl)pyrrolo[3,2-b]pyridine-3-carboxamide | | 著者 | Chen, X.J, Lin, Q.M, Chen, Y.H. | | 登録日 | 2022-06-29 | | 公開日 | 2022-11-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.243 Å) | | 主引用文献 | Design, Synthesis, and Biological Evaluation of 5-Formyl-pyrrolo[3,2- b ]pyridine-3-carboxamides as New Selective, Potent, and Reversible-Covalent FGFR4 Inhibitors.
J.Med.Chem., 65, 2022
|
|
