8IIO
 
 | | H109Q mutant of uracil DNA glycosylase X | | 分子名称: | GLYCEROL, IRON/SULFUR CLUSTER, Type-4 uracil-DNA glycosylase | | 著者 | Aroli, S. | | 登録日 | 2023-02-24 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Mutational and structural analyses of UdgX: insights into the active site pocket architecture and its evolution.
Nucleic Acids Res., 51, 2023
|
|
8IIM
 
 | | H109K mutant of uracil DNA glycosylase X | | 分子名称: | BETA-MERCAPTOETHANOL, IRON/SULFUR CLUSTER, Type-4 uracil-DNA glycosylase | | 著者 | Aroli, S. | | 登録日 | 2023-02-24 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Mutational and structural analyses of UdgX: insights into the active site pocket architecture and its evolution.
Nucleic Acids Res., 51, 2023
|
|
8IIT
 
 | |
7VUF
 
 | |
8IIL
 
 | |
8IIF
 
 | | H109A mutant of uracil DNA glycosylase X | | 分子名称: | GLYCEROL, IRON/SULFUR CLUSTER, Type-4 uracil-DNA glycosylase | | 著者 | Aroli, S. | | 登録日 | 2023-02-24 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Mutational and structural analyses of UdgX: insights into the active site pocket architecture and its evolution.
Nucleic Acids Res., 51, 2023
|
|
8IIH
 
 | | H109C mutant of uracil DNA glycosylase X | | 分子名称: | IRON/SULFUR CLUSTER, Type-4 uracil-DNA glycosylase | | 著者 | Aroli, S. | | 登録日 | 2023-02-24 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Mutational and structural analyses of UdgX: insights into the active site pocket architecture and its evolution.
Nucleic Acids Res., 51, 2023
|
|
8IIJ
 
 | | H109G mutant of uracil DNA glycosylase X | | 分子名称: | BETA-MERCAPTOETHANOL, IRON/SULFUR CLUSTER, Type-4 uracil-DNA glycosylase | | 著者 | Aroli, S. | | 登録日 | 2023-02-24 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Mutational and structural analyses of UdgX: insights into the active site pocket architecture and its evolution.
Nucleic Acids Res., 51, 2023
|
|
7VUK
 
 | |
5K8J
 
 | | Structure of Caulobacter crescentus VapBC1 (apo form) | | 分子名称: | GLYCEROL, Ribonuclease VapC, VapB family protein | | 著者 | Bendtsen, K.L, Xu, K, Luckmann, M, Brodersen, D.E. | | 登録日 | 2016-05-30 | | 公開日 | 2016-12-28 | | 最終更新日 | 2018-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Toxin inhibition in C. crescentus VapBC1 is mediated by a flexible pseudo-palindromic protein motif and modulated by DNA binding.
Nucleic Acids Res., 45, 2017
|
|
7KPO
 
 | | High Resolution Crystal Structure of the DNA-binding Domain from the Sensor Histidine Kinase ChiS from Vibrio cholerae | | 分子名称: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, Response regulator, ... | | 著者 | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-11-12 | | 公開日 | 2020-11-25 | | 実験手法 | X-RAY DIFFRACTION (1.28 Å) | | 主引用文献 | High Resolution Crystal Structure of the DNA-binding Domain from the Sensor Histidine Kinase ChiS from Vibrio cholerae.
To Be Published
|
|
5VMW
 
 | | Kaiso (ZBTB33) zinc finger DNA binding domain in complex with a double CpG-methylated DNA resembling the specific Kaiso binding sequence (KBS) | | 分子名称: | CHLORIDE ION, DNA (5'-D(*CP*GP*TP*TP*AP*TP*TP*(5CM)P*GP*(5CM)P*GP*GP*GP*AP*AP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*TP*CP*CP*(5CM)P*GP*(5CM)P*GP*AP*AP*TP*AP*AP*CP*G)-3'), ... | | 著者 | Nikolova, E.N, Stanfield, R.L, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | 登録日 | 2017-04-28 | | 公開日 | 2018-04-04 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.397 Å) | | 主引用文献 | CH···O Hydrogen Bonds Mediate Highly Specific Recognition of Methylated CpG Sites by the Zinc Finger Protein Kaiso.
Biochemistry, 57, 2018
|
|
7L6L
 
 | | Crystal Structure of the DNA-binding Transcriptional Repressor DeoR from Escherichia coli str. K-12 | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Deoxyribose operon repressor, ... | | 著者 | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-12-23 | | 公開日 | 2021-12-01 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal Structure of the DNA-binding Transcriptional Repressor DeoR from Escherichia coli str. K-12.
To Be Published
|
|
5L6M
 
 | | Structure of Caulobacter crescentus VapBC1 (VapB1deltaC:VapC1 form) | | 分子名称: | GLYCEROL, MALONATE ION, Ribonuclease VapC, ... | | 著者 | Bendtsen, K.L, Xu, K, Luckmann, M, Brodersen, D.E. | | 登録日 | 2016-05-30 | | 公開日 | 2016-12-28 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Toxin inhibition in C. crescentus VapBC1 is mediated by a flexible pseudo-palindromic protein motif and modulated by DNA binding.
Nucleic Acids Res., 45, 2017
|
|
4K1G
 
 | |
3FT7
 
 | | Crystal structure of an extremely stable dimeric protein from sulfolobus islandicus | | 分子名称: | GLYCEROL, Uncharacterized protein ORF56 | | 著者 | Neumann, P, Loew, C, Weininger, U, Stubbs, M.T. | | 登録日 | 2009-01-12 | | 公開日 | 2009-10-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-Based Stability Analysis of an Extremely Stable Dimeric DNA Binding Protein from Sulfolobus islandicus
Biochemistry, 48, 2009
|
|
4N6R
 
 | | Crystal structure of VosA-VelB-complex | | 分子名称: | SULFATE ION, VelB, VosA | | 著者 | Ahmed, Y.L, Dickmanns, A, Neumann, P, Ficner, R. | | 登録日 | 2013-10-14 | | 公開日 | 2014-01-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The Velvet Family of Fungal Regulators Contains a DNA-Binding Domain Structurally Similar to NF-kappa B.
Plos Biol., 11, 2013
|
|
4M34
 
 | | Crystal structure of gated-pore mutant D138H of second DNA-Binding protein under starvation from Mycobacterium smegmatis | | 分子名称: | CHLORIDE ION, FE (II) ION, MAGNESIUM ION, ... | | 著者 | Williams, S.M, Chandran, A.V, Vijayabaskar, M.S, Roy, S, Balaram, H, Vishveshwara, S, Vijayan, M, Chatterji, D. | | 登録日 | 2013-08-06 | | 公開日 | 2014-03-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | A histidine aspartate ionic lock gates the iron passage in miniferritins from Mycobacterium smegmatis
J.Biol.Chem., 289, 2014
|
|
3ZQM
 
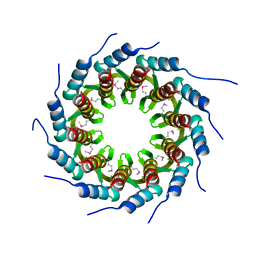 | | Crystal structure of the small terminase oligomerization core domain from a SPP1-like bacteriophage (crystal form 1) | | 分子名称: | TERMINASE SMALL SUBUNIT | | 著者 | Buttner, C.R, Chechik, M, Ortiz-Lombardia, M, Smits, C, Chechik, V, Jeschke, G, Dykeman, E, Benini, S, Alonso, J.C, Antson, A.A. | | 登録日 | 2011-06-10 | | 公開日 | 2011-12-28 | | 最終更新日 | 2012-02-15 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural Basis for DNA Recognition and Loading Into a Viral Packaging Motor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1QPS
 
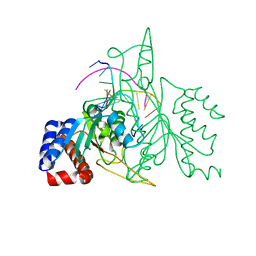 | | THE CRYSTAL STRUCTURE OF A POST-REACTIVE COGNATE DNA-ECO RI COMPLEX AT 2.50 A IN THE PRESENCE OF MN2+ ION | | 分子名称: | 5'-D(*AP*AP*TP*TP*CP*GP*CP*GP*)-3', 5'-D(*TP*CP*GP*CP*GP*)-3', ENDONUCLEASE ECORI, ... | | 著者 | Horvath, M, Choi, J, Kim, Y, Wilkosz, P, Rosenberg, J.M. | | 登録日 | 1999-05-28 | | 公開日 | 1999-06-14 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The Integration of Recognition and Cleavage: X-Ray Structures of Pre- Transition State and Post-Reactive DNA-Eco RI Endonuclease Complexes
To be Published
|
|
1F6O
 
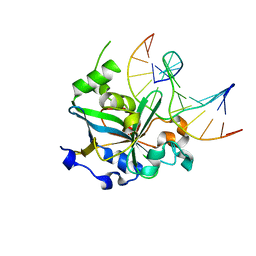 | | CRYSTAL STRUCTURE OF THE HUMAN AAG DNA REPAIR GLYCOSYLASE COMPLEXED WITH DNA | | 分子名称: | 3-METHYL-ADENINE DNA GLYCOSYLASE, DNA (5'-D(*GP*AP*CP*AP*TP*GP*(YRR)P*TP*TP*GP*CP*CP*T)-3'), DNA (5'-D(*GP*GP*CP*AP*AP*TP*CP*AP*TP*GP*TP*CP*A)-3'), ... | | 著者 | Lau, A.Y, Wyatt, M.D, Glassner, B.J, Samson, L.D, Ellenberger, T. | | 登録日 | 2000-06-22 | | 公開日 | 2000-12-11 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Molecular basis for discriminating between normal and damaged bases by the human alkyladenine glycosylase, AAG.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
2L31
 
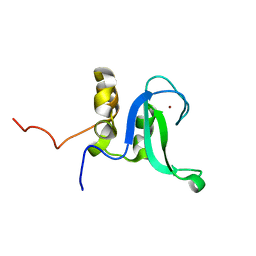 | | Human PARP-1 zinc finger 2 | | 分子名称: | Poly [ADP-ribose] polymerase 1, ZINC ION | | 著者 | Neuhaus, D, Eustermann, S, Yang, J, Videler, H. | | 登録日 | 2010-08-30 | | 公開日 | 2011-02-02 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The DNA-binding domain of human PARP-1 interacts with DNA single-strand breaks as a monomer through its second zinc finger.
J.Mol.Biol., 407, 2011
|
|
2L30
 
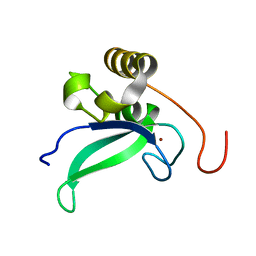 | | Human PARP-1 zinc finger 1 | | 分子名称: | Poly [ADP-ribose] polymerase 1, ZINC ION | | 著者 | Neuhaus, D, Eustermann, S, Yang, J, Videler, H. | | 登録日 | 2010-08-30 | | 公開日 | 2011-02-02 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The DNA-binding domain of human PARP-1 interacts with DNA single-strand breaks as a monomer through its second zinc finger.
J.Mol.Biol., 407, 2011
|
|
2F8W
 
 | | Crystal structure of d(CACGTG)2 | | 分子名称: | 1,3-DIAMINOPROPANE, 5'-D(*CP*AP*CP*GP*TP*G)-3', SPERMINE | | 著者 | Narayana, N, Shamala, N, Ganesh, K.N, Viswamitra, M.A. | | 登録日 | 2005-12-04 | | 公開日 | 2006-01-31 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Interaction between the Z-Type DNA Duplex and 1,3-Propanediamine: Crystal Structure of d(CACGTG)2 at 1.2 A Resolution
Biochemistry, 45, 2006
|
|
5U34
 
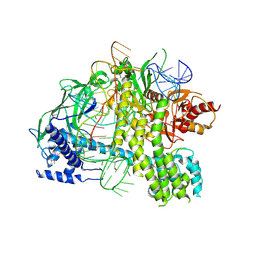 | | Crystal structure of AacC2c1-sgRNA binary complex | | 分子名称: | CRISPR-associated endonuclease C2c1, sgRNA | | 著者 | Yang, H, Gao, P, Rajashankar, K.R, Patel, D.J. | | 登録日 | 2016-12-01 | | 公開日 | 2017-01-25 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.255 Å) | | 主引用文献 | PAM-Dependent Target DNA Recognition and Cleavage by C2c1 CRISPR-Cas Endonuclease.
Cell, 167, 2016
|
|
