4L8Q
 
 | | Crystal structure of Canavalia grandiflora seed lectin complexed with X-Man. | | 分子名称: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CADMIUM ION, CALCIUM ION, ... | | 著者 | Barroso-Neto, I.L, Rocha, B.A.M, Simoes, R.C, Bezerra, M.J.B, Pereira-Junior, F.N, Osterne, V.J.S, Nascimento, K.S, Nagano, C.S, Delatorre, P, Sampaio, A.H, Cavada, B.S. | | 登録日 | 2013-06-17 | | 公開日 | 2014-05-21 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Vasorelaxant activity of Canavalia grandiflora seed lectin: A structural analysis.
Arch.Biochem.Biophys., 543, 2014
|
|
3U95
 
 | | Crystal structure of a putative alpha-glucosidase from Thermotoga neapolitana | | 分子名称: | Glycoside hydrolase, family 4, MANGANESE (II) ION | | 著者 | Ha, N.C, Jun, S.Y, Yun, B.Y, Yoon, B.Y, Piao, S. | | 登録日 | 2011-10-17 | | 公開日 | 2012-09-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.998 Å) | | 主引用文献 | Crystal structure and thermostability of a putative alpha-glucosidase from Thermotoga neapolitana
Biochem.Biophys.Res.Commun., 416, 2011
|
|
3OY1
 
 | | Highly Selective c-Jun N-Terminal Kinase (JNK) 2 and 3 Inhibitors with In Vitro CNS-like Pharmacokinetic Properties | | 分子名称: | 5-[2-(cyclohexylamino)pyridin-4-yl]-4-naphthalen-2-yl-2-(tetrahydro-2H-pyran-4-yl)-2,4-dihydro-3H-1,2,4-triazol-3-one, Mitogen-activated protein kinase 10 | | 著者 | Probst, G.D, Bowers, S, Sealy, J.M, Truong, A, Neitz, J, Hom, R.K, Galemmo Jr, R.A, Konradi, A.W, Sham, H.L, Quincy, D, Pan, H, Yao, N. | | 登録日 | 2010-09-22 | | 公開日 | 2011-08-17 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Highly selective c-Jun N-terminal kinase (JNK) 2 and 3 inhibitors with in vitro CNS-like pharmacokinetic properties prevent neurodegeneration.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2QNF
 
 | | Crystal structure of T4 Endonuclease VII H43N mutant in complex with heteroduplex DNA containing base mismatches | | 分子名称: | DNA (5'-D(*DCP*DAP*DCP*DAP*DTP*DCP*DGP*DAP*DTP*DGP*DGP*DAP*DGP*DCP*DCP*DG)-3'), DNA (5'-D(*DCP*DAP*DCP*DAP*DTP*DCP*DGP*DAP*DTP*DGP*DGP*DAP*DGP*DCP*DGP*DC)-3'), DNA (5'-D(*DCP*DGP*DGP*DCP*DTP*DCP*DCP*DAP*DTP*DCP*DGP*DAP*DTP*DGP*DTP*DG)-3'), ... | | 著者 | Biertumpfel, C, Yang, W, Suck, D. | | 登録日 | 2007-07-18 | | 公開日 | 2008-01-29 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structure of T4 endonuclease VII resolving a Holliday junction.
Nature, 449, 2007
|
|
1DCV
 
 | |
1CUK
 
 | |
7UTJ
 
 | | Cryogenic electron microscopy 3D map of F-actin bound by human dimeric alpha-catenin | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | 著者 | Rangarajan, E.S, Smith, E.W, Izard, T. | | 登録日 | 2022-04-27 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-03-29 | | 実験手法 | ELECTRON MICROSCOPY (2.77 Å) | | 主引用文献 | Distinct inter-domain interactions of dimeric versus monomeric alpha-catenin link cell junctions to filaments.
Commun Biol, 6, 2023
|
|
3PGD
 
 | | Crystal Structure of HLA-DR1 with CLIP106-120, canonical peptide orientation | | 分子名称: | HLA class II histocompatibility antigen gamma chain, HLA class II histocompatibility antigen, DR alpha chain, ... | | 著者 | Gunther, S, Schlundt, A, Sticht, J, Roske, Y, Heinemann, U, Wiesmuller, K.-H, Jung, G, Falk, K, Rotzschke, O, Freund, C. | | 登録日 | 2010-11-01 | | 公開日 | 2010-12-08 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.72 Å) | | 主引用文献 | Bidirectional binding of invariant chain peptides to an MHC class II molecule.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1M0I
 
 | | Crystal Structure of Bacteriophage T7 Endonuclease I with a Wild-Type Active Site | | 分子名称: | SULFATE ION, endodeoxyribonuclease I | | 著者 | Hadden, J.M, Declais, A.C, Phillips, S.E, Lilley, D.M. | | 登録日 | 2002-06-13 | | 公開日 | 2002-12-18 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Metal ions bound at the active site of the junction-resolving enzyme T7 endonuclease I
Embo J., 21, 2002
|
|
7MFW
 
 | |
7Z6H
 
 | | Structure of DNA-bound human RAD17-RFC clamp loader and 9-1-1 checkpoint clamp | | 分子名称: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1,Cell cycle checkpoint protein RAD17, Checkpoint protein HUS1, ... | | 著者 | Day, M, Oliver, A.W, Pearl, L.H. | | 登録日 | 2022-03-11 | | 公開日 | 2022-05-04 | | 最終更新日 | 2022-08-31 | | 実験手法 | ELECTRON MICROSCOPY (3.59 Å) | | 主引用文献 | Structure of the human RAD17-RFC clamp loader and 9-1-1 checkpoint clamp bound to a dsDNA-ssDNA junction.
Nucleic Acids Res., 50, 2022
|
|
8ITS
 
 | |
7VUF
 
 | |
7VUK
 
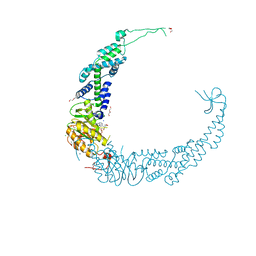 | |
1D3R
 
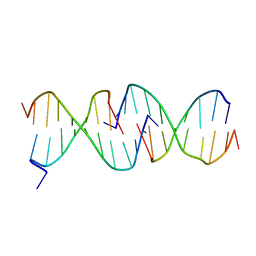 | | CRYSTAL STRUCTURE OF TRIPLEX DNA | | 分子名称: | DNA (5'-D(*CP*(BRU)P*CP*CP*(BRU)P*CP*CP*GP*CP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*GP*CP*GP*GP*AP*G)-3') | | 著者 | Rhee, S, Han, Z.-J, Liu, K, Todd Miles, H.T, Davies, D.R. | | 登録日 | 1999-09-30 | | 公開日 | 2000-01-01 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of a triple helical DNA with a triplex-duplex junction.
Biochemistry, 38, 1999
|
|
8ABD
 
 | |
8ABN
 
 | |
1EN7
 
 | | ENDONUCLEASE VII (ENDOVII) FROM PHAGE T4 | | 分子名称: | CALCIUM ION, RECOMBINATION ENDONUCLEASE VII, ZINC ION | | 著者 | Raaijmakers, H, Vix, O, Toro, I, Suck, D. | | 登録日 | 1999-02-07 | | 公開日 | 2000-02-07 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | X-ray structure of T4 endonuclease VII: a DNA junction resolvase with a novel fold and unusual domain-swapped dimer architecture.
EMBO J., 18, 1999
|
|
4K1N
 
 | |
6D57
 
 | | Campylobacter jejuni ferric uptake regulator S1 metalated | | 分子名称: | FORMIC ACID, Ferric uptake regulation protein, GLYCEROL, ... | | 著者 | Sarvan, S, Brunzelle, J.S, Couture, J.F. | | 登録日 | 2018-04-19 | | 公開日 | 2018-05-23 | | 最終更新日 | 2020-01-08 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Functional insights into the interplay between DNA interaction and metal coordination in ferric uptake regulators.
Sci Rep, 8, 2018
|
|
5A5D
 
 | | A complex of the synthetic siderophore analogue Fe(III)-5-LICAM with the CeuE periplasmic protein from Campylobacter jejuni | | 分子名称: | ENTEROCHELIN UPTAKE PERIPLASMIC BINDING PROTEIN, FE (III) ION, N,N'-pentane-1,5-diylbis(2,3-dihydroxybenzamide) | | 著者 | Blagova, E, Hughes, A, Moroz, O.V, Raines, D.J, Wilde, E.J, Turkenburg, J.P, Duhme-Klair, A.-K, Wilson, K.S. | | 登録日 | 2015-06-17 | | 公開日 | 2016-06-29 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Interactions of the periplasmic binding protein CeuE with Fe(III) n-LICAM(4-) siderophore analogues of varied linker length.
Sci Rep, 7, 2017
|
|
5AD1
 
 | | A complex of the synthetic siderophore analogue Fe(III)-8-LICAM with the CeuE periplasmic protein from Campylobacter jejuni | | 分子名称: | ENTEROCHELIN UPTAKE PERIPLASMIC BINDING PROTEIN, FE (III) ION, N,N'-OCTANE-1,8-DIYLBIS(2,3-DIHYDROXYBENZAMIDE) | | 著者 | Blagova, E, Hughes, A, Moroz, O.V, Raines, D.J, Wilde, E.J, Turkenburg, J.P, Duhme-Klair, A.-K, Wilson, K.S. | | 登録日 | 2015-08-19 | | 公開日 | 2016-09-28 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | Interactions of the periplasmic binding protein CeuE with Fe(III) n-LICAM(4-) siderophore analogues of varied linker length.
Sci Rep, 7, 2017
|
|
6UVT
 
 | |
1KPD
 
 | |
2MI0
 
 | |
