5ZCE
 
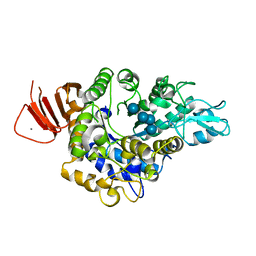 | | Crystal structure of Alpha-glucosidase in complex with maltotetraose | | 分子名称: | Alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Kato, K, Saburi, W, Yao, M. | | 登録日 | 2018-02-16 | | 公開日 | 2018-12-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.555 Å) | | 主引用文献 | Function and structure of GH13_31 alpha-glucosidase with high alpha-(1→4)-glucosidic linkage specificity and transglucosylation activity.
FEBS Lett., 592, 2018
|
|
6ANV
 
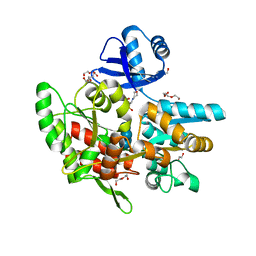 | | Crystal structure of anti-CRISPR protein AcrF1 | | 分子名称: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Yang, H, Patel, D.J. | | 登録日 | 2017-08-14 | | 公開日 | 2017-10-25 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.265 Å) | | 主引用文献 | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
6B3P
 
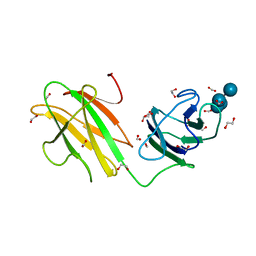 | | Crystal structure of CBMbc (family CBM26) from Eubacterium rectale Amy13K in Complex with Maltoheptaose | | 分子名称: | 1,2-ETHANEDIOL, Amy13K, FORMIC ACID, ... | | 著者 | Cockburn, D.W, Wawrzak, Z, Perez Medina, K, Koropatkin, N.M. | | 登録日 | 2017-09-22 | | 公開日 | 2017-11-29 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Novel carbohydrate binding modules in the surface anchored alpha-amylase of Eubacterium rectale provide a molecular rationale for the range of starches used by this organism in the human gut.
Mol. Microbiol., 107, 2018
|
|
6BS6
 
 | |
6DTS
 
 | | Maltotetraose bound T. maritima MalE2 | | 分子名称: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltose-binding protein MalE2 | | 著者 | Cuneo, M.J, Shukla, S. | | 登録日 | 2018-06-18 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Differential Substrate Recognition by Maltose Binding Proteins Influenced by Structure and Dynamics.
Biochemistry, 57, 2018
|
|
6DTU
 
 | | Maltotetraose bound T. maritima MalE1 | | 分子名称: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltose-binding protein MalE1 | | 著者 | Cuneo, M.J, Shukla, S. | | 登録日 | 2018-06-18 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Differential Substrate Recognition by Maltose Binding Proteins Influenced by Structure and Dynamics.
Biochemistry, 57, 2018
|
|
6IYG
 
 | | The Structure of Maltooligosaccharide-forming Amylase from Pseudomonas saccharophila STB07 with Maltotetraose | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, Glucan 1,4-alpha-maltotetraohydrolase, ... | | 著者 | Li, Z.F, Ban, X.F, Zhang, Z.Q, Li, C.M, Gu, Z.B, Jin, T.C, Li, Y.L, Shang, Y.H. | | 登録日 | 2018-12-15 | | 公開日 | 2019-12-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Maltotetraose-forming amylase from Pseudomonas saccharophila STB07
To Be Published
|
|
6L3E
 
 | | Crystal structure of Salmonella enterica sugar-binding protein MalE | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | 著者 | Wang, L, Chen, Y, Liu, W, Lan, J, Shang, F, Xu, Y. | | 登録日 | 2019-10-10 | | 公開日 | 2019-10-23 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The crystal structure of Salmonella enterica sugar-binding protein MalE
To Be Published
|
|
6M4M
 
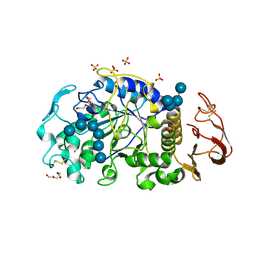 | | X-ray crystal structure of the E249Q mutan of alpha-amylase I and maltohexaose complex from Eisenia fetida | | 分子名称: | Alpha-amylase, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Hirano, Y, Tsukamoto, K, Ariki, S, Naka, Y, Ueda, M, Tamada, T. | | 登録日 | 2020-03-07 | | 公開日 | 2020-09-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | X-ray crystallographic structural studies of alpha-amylase I from Eisenia fetida.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6SMV
 
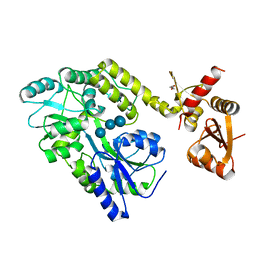 | | Structure of HPV49 E6 protein in complex with MAML1 LxxLL motif | | 分子名称: | DI(HYDROXYETHYL)ETHER, Maltose/maltodextrin-binding periplasmic protein,Protein E6,Mastermind-like protein 1, ZINC ION, ... | | 著者 | Suarez, I.P, Cousido-Siah, A, Bonhoure, A, Kostmann, C, Mitschler, A, Podjarny, A, Trave, G. | | 登録日 | 2019-08-22 | | 公開日 | 2019-09-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Cellular target recognition by HPV18 and HPV49 oncoproteins
To be published
|
|
6TP1
 
 | |
7ML5
 
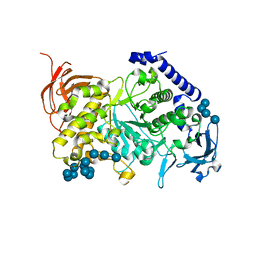 | | Structure of the Starch Branching Enzyme I (BEI) complexed with maltododecaose from Oryza sativa L | | 分子名称: | Isoform 2 of 1,4-alpha-glucan-branching enzyme, chloroplastic/amyloplastic, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | 著者 | Nayebi Gavgani, H, Fawaz, R, Geiger, J.H. | | 登録日 | 2021-04-27 | | 公開日 | 2021-11-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | A structural explanation for the mechanism and specificity of plant branching enzymes I and IIb.
J.Biol.Chem., 298, 2021
|
|
8DHD
 
 | | Neutron crystal structure of maltotetraose bound tmMBP | | 分子名称: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltose-binding protein MalE2 | | 著者 | Cuneo, M.J, Shukla, S, Myles, D.A. | | 登録日 | 2022-06-27 | | 公開日 | 2022-10-12 | | 最終更新日 | 2023-10-25 | | 実験手法 | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | 主引用文献 | Mapping periplasmic binding protein oligosaccharide recognition with neutron crystallography.
Sci Rep, 12, 2022
|
|
8DL1
 
 | | BoGH13ASus-E523Q from Bacteroides ovatus bound to maltoheptaose | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, Alpha amylase, ... | | 著者 | Brown, H.A, DeVeaux, A.L, Koropatkin, N.M. | | 登録日 | 2022-07-06 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | BoGH13A Sus from Bacteroides ovatus represents a novel alpha-amylase used for Bacteroides starch breakdown in the human gut.
Cell.Mol.Life Sci., 80, 2023
|
|
8H68
 
 | | Crystal structure of Caenorhabditis elegans NMAD-1 in complex with NOG and Mg(II) | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Shi, Y, Ding, J, Yang, H. | | 登録日 | 2022-10-16 | | 公開日 | 2023-02-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Caenorhabditis elegans NMAD-1 functions as a demethylase for actin.
J Mol Cell Biol, 15, 2023
|
|
8IIY
 
 | |
8SDB
 
 | | Crystal Structure of E.Coli Branching Enzyme in complex with malto-octose | | 分子名称: | 1,4-alpha-glucan branching enzyme GlgB, alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | 著者 | Bingham, C.R, Nayebi, H, Fawaz, R, Geiger, J.H. | | 登録日 | 2023-04-06 | | 公開日 | 2023-07-12 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The Structure of Maltooctaose-Bound Escherichia coli Branching Enzyme Suggests a Mechanism for Donor Chain Specificity.
Molecules, 28, 2023
|
|
