2WLX
 
 | |
3OP3
 
 | | Crystal Structure of Cell Division Cycle 25C Protein Isoform A from Homo sapiens | | 分子名称: | M-phase inducer phosphatase 3, SULFATE ION | | 著者 | Kim, Y, Weger, A, Hatzos, C, Savitsky, P, Johansson, C, Ball, L, Barr, A, Vollmar, M, Muniz, J, Weigelt, J, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O, von Delft, F, Knapp, S, Joachimiak, A, Structural Genomics Consortium (SGC) | | 登録日 | 2010-08-31 | | 公開日 | 2010-09-29 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.63 Å) | | 主引用文献 | Crystal Structure of Cell Division Cycle 25C Protein Isoform A from Homo sapiens
TO BE PUBLISHED
|
|
3OLH
 
 | | Human 3-mercaptopyruvate sulfurtransferase | | 分子名称: | 3-mercaptopyruvate sulfurtransferase, SODIUM ION, SULFATE ION | | 著者 | Karlberg, T, Collins, R, Arrowsmith, C.H, Berglund, H, Bountra, C, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kotenyova, T, Kouznetsova, E, Moche, M, Nordlund, P, Nyman, T, Persson, C, Schutz, P, Sehic, A, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | 登録日 | 2010-08-26 | | 公開日 | 2010-09-29 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Human 3-Mercaptopyruvate Sulfurtransferase
To be Published
|
|
3P3A
 
 | |
3NTA
 
 | | Structure of the Shewanella loihica PV-4 NADH-dependent persulfide reductase | | 分子名称: | CHLORIDE ION, COENZYME A, FAD-dependent pyridine nucleotide-disulphide oxidoreductase, ... | | 著者 | Sazinsky, M.H, Crane, E.J, Warner, M.D, Lukose, V, Lee, K.H. | | 登録日 | 2010-07-03 | | 公開日 | 2010-12-08 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Characterization of an NADH-Dependent Persulfide Reductase from Shewanella loihica PV-4: Implications for the Mechanism of Sulfur Respiration via FAD-Dependent Enzymes .
Biochemistry, 50, 2010
|
|
3NT6
 
 | | Structure of the Shewanella loihica PV-4 NADH-dependent persulfide reductase C43S/C531S Double Mutant | | 分子名称: | CHLORIDE ION, COENZYME A, FAD-dependent pyridine nucleotide-disulphide oxidoreductase, ... | | 著者 | Sazinsky, M.H, Crane, E.J, Warner, M.D, Lukose, V, Lee, K.H, Lopez, K. | | 登録日 | 2010-07-02 | | 公開日 | 2010-12-08 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Characterization of an NADH-Dependent Persulfide Reductase from Shewanella loihica PV-4: Implications for the Mechanism of Sulfur Respiration via FAD-Dependent Enzymes .
Biochemistry, 50, 2010
|
|
3NTD
 
 | | Structure of the Shewanella loihica PV-4 NADH-dependent persulfide reductase C531S Mutant | | 分子名称: | CHLORIDE ION, COENZYME A, FAD-dependent pyridine nucleotide-disulphide oxidoreductase, ... | | 著者 | Sazinsky, M.H, Warner, M.D, Lukose, V, Lee, K.H, Crane, E.J. | | 登録日 | 2010-07-03 | | 公開日 | 2010-12-08 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Characterization of an NADH-Dependent Persulfide Reductase from Shewanella loihica PV-4: Implications for the Mechanism of Sulfur Respiration via FAD-Dependent Enzymes .
Biochemistry, 50, 2010
|
|
3R2U
 
 | | 2.1 Angstrom Resolution Crystal Structure of Metallo-beta-lactamase from Staphylococcus aureus subsp. aureus COL | | 分子名称: | CHLORIDE ION, FE (III) ION, MAGNESIUM ION, ... | | 著者 | Minasov, G, Wawrzak, Z, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Kiryukhina, O, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-03-14 | | 公開日 | 2011-03-23 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | 2.1 Angstrom Resolution Crystal Structure of Metallo-beta-lactamase Family Protein from Staphylococcus aureus subsp. aureus COL
TO BE PUBLISHED
|
|
3TP9
 
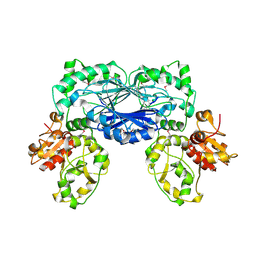 | | Crystal structure of Alicyclobacillus acidocaldarius protein with beta-lactamase and rhodanese domains | | 分子名称: | BETA-LACTAMASE and RHODANESE DOMAIN PROTEIN, ZINC ION | | 著者 | Michalska, K, Chhor, G, Mandel, M.E, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-09-07 | | 公開日 | 2011-09-21 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of Alicyclobacillus acidocaldarius protein with beta-lactamase and rhodanese domains
To be Published
|
|
3TG1
 
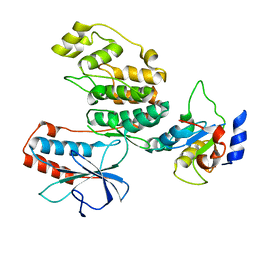 | |
3TG3
 
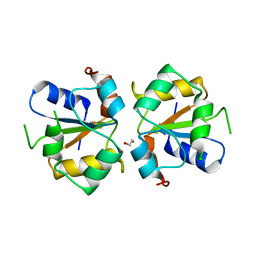 | | Crystal structure of the MAPK binding domain of MKP7 | | 分子名称: | 1,2-ETHANEDIOL, Dual specificity protein phosphatase 16 | | 著者 | Zhang, Y.Y, Liu, X, Wu, J.W, Wang, Z.X. | | 登録日 | 2011-08-17 | | 公開日 | 2012-03-14 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.675 Å) | | 主引用文献 | A Distinct Interaction Mode Revealed by the Crystal Structure of the Kinase p38alpha with the MAPK Binding Domain of the Phosphatase MKP5.
Sci.Signal., 4, 2011
|
|
4F67
 
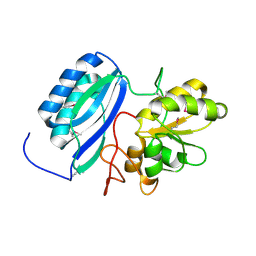 | | Three dimensional structure of the double mutant of UPF0176 protein lpg2838 from Legionella pneumophila at the resolution 1.8A, Northeast Structural Genomics Consortium (NESG) Target LgR82 | | 分子名称: | UPF0176 protein lpg2838 | | 著者 | Kuzin, A, Neely, H, Street, L, Odukwe, N, Seetharaman, J, Mao, M, Xiao, R, Kohan, E, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2012-05-14 | | 公開日 | 2012-05-30 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Three dimensional structure of the double mutant of UPF0176 protein lpg2838 from Legionella pneumophila at the resolution 1.8A, Northeast
Structural Genomics Consortium (NESG) Target LgR82
To be Published
|
|
3UTN
 
 | | Crystal structure of Tum1 protein from Saccharomyces cerevisiae | | 分子名称: | DIMETHYL SULFOXIDE, SULFATE ION, Thiosulfate sulfurtransferase TUM1 | | 著者 | Qiu, R, Wang, F, Liu, M, Ji, C, Gong, W. | | 登録日 | 2011-11-26 | | 公開日 | 2012-10-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of the Tum1 protein from the yeast Saccharomyces cerevisiae.
Protein Pept.Lett., 19, 2012
|
|
4JGT
 
 | | Structure and kinetic analysis of H2S production by human Mercaptopyruvate Sulfurtransferase | | 分子名称: | 3-mercaptopyruvate sulfurtransferase, GLYCEROL, PYRUVIC ACID, ... | | 著者 | Koutmos, M, Yamada, K, Yadav, P.K, Chiku, T, Banerjee, R. | | 登録日 | 2013-03-03 | | 公開日 | 2013-05-29 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.161 Å) | | 主引用文献 | Structure and Kinetic Analysis of H2S Production by Human Mercaptopyruvate Sulfurtransferase.
J.Biol.Chem., 288, 2013
|
|
2MOI
 
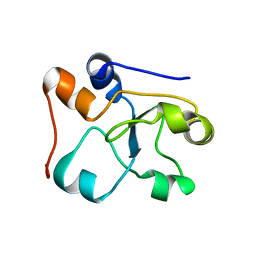 | | 3D NMR structure of the cytoplasmic rhodanese domain of the inner membrane protein YgaP from Escherichia coli | | 分子名称: | Inner membrane protein YgaP | | 著者 | Eichmann, C, Tzitzilonis, C, Bordignon, E, Maslennikov, I, Choe, S, Riek, R. | | 登録日 | 2014-04-26 | | 公開日 | 2014-06-25 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR Structure and Functional Analysis of the Integral Membrane Protein YgaP from Escherichia coli.
J.Biol.Chem., 289, 2014
|
|
2MOL
 
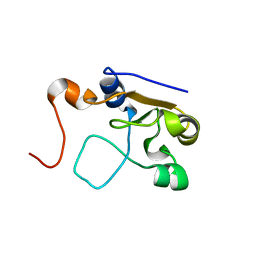 | | 3D NMR structure of the cytoplasmic rhodanese domain of the full-length inner membrane protein YgaP from Escherichia coli | | 分子名称: | Inner membrane protein YgaP | | 著者 | Eichmann, C, Tzitzilonis, C, Bordignon, E, Maslennikov, I, Choe, S, Riek, R. | | 登録日 | 2014-04-27 | | 公開日 | 2014-06-25 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR Structure and Functional Analysis of the Integral Membrane Protein YgaP from Escherichia coli.
J.Biol.Chem., 289, 2014
|
|
4OCG
 
 | |
4WH9
 
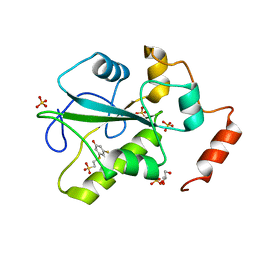 | | Structure of the CDC25B Phosphatase Catalytic Domain with Bound Inhibitor | | 分子名称: | 2-[(2-cyano-3-fluoro-5-hydroxyphenyl)sulfanyl]ethanesulfonic acid, GLYCEROL, M-phase inducer phosphatase 2, ... | | 著者 | Lund, G.L, Dudkin, S, Borkin, D, Ni, W, Grembecka, J, Cierpicki, T. | | 登録日 | 2014-09-20 | | 公開日 | 2014-12-10 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Inhibition of CDC25B Phosphatase Through Disruption of Protein-Protein Interaction.
Acs Chem.Biol., 10, 2015
|
|
4WH7
 
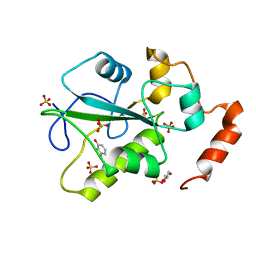 | | Structure of the CDC25B Phosphatase Catalytic Domain with Bound Ligand | | 分子名称: | 2-fluoro-4-hydroxybenzonitrile, GLYCEROL, M-phase inducer phosphatase 2, ... | | 著者 | Lund, G.L, Dudkin, S, Borkin, D, Ni, W, Grembecka, J, Cierpicki, T. | | 登録日 | 2014-09-20 | | 公開日 | 2014-12-10 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Inhibition of CDC25B Phosphatase Through Disruption of Protein-Protein Interaction.
Acs Chem.Biol., 10, 2015
|
|
5HBO
 
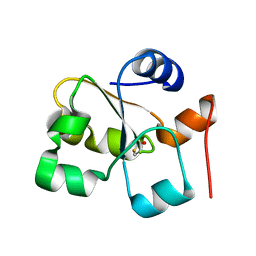 | | Native rhodanese domain of YgaP prepared without DDT is both S-nitrosylated and S-sulfhydrated | | 分子名称: | Inner membrane protein YgaP | | 著者 | Eichmann, C, Tzitzilonis, C, Nakamura, T, Kwiatkowski, W, Maslennikov, I, Choe, S, Lipton, S.A, Riek, R. | | 登録日 | 2016-01-01 | | 公開日 | 2016-08-10 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
5HBL
 
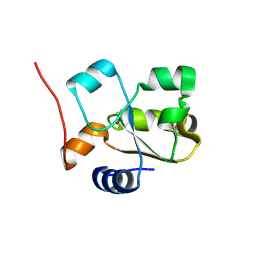 | | Native rhodanese domain of YgaP prepared with 1mM DDT is S-nitrosylated | | 分子名称: | Inner membrane protein YgaP | | 著者 | Eichmann, C, Tzitzilonis, C, Nakamura, T, Kwiatkowski, W, Maslennikov, I, Choe, S, Lipton, S.A, Riek, R. | | 登録日 | 2015-12-31 | | 公開日 | 2016-08-10 | | 最終更新日 | 2021-09-08 | | 実験手法 | X-RAY DIFFRACTION (1.617 Å) | | 主引用文献 | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
5HBQ
 
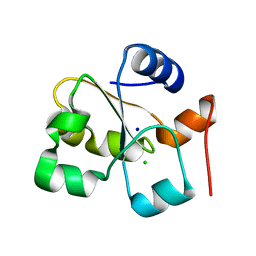 | | C63D mutant of the rhodanese domain of YgaP | | 分子名称: | CHLORIDE ION, Inner membrane protein YgaP, SODIUM ION | | 著者 | Eichmann, C, Tzitzilonis, C, Nakamura, T, Kwiatkowski, W, Maslennikov, I, Choe, S, Lipton, S.A, Riek, R. | | 登録日 | 2016-01-02 | | 公開日 | 2016-08-10 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
5HBP
 
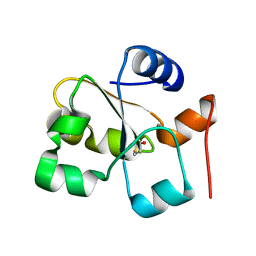 | | The crystal of rhodanese domain of YgaP treated with SNOC | | 分子名称: | Inner membrane protein YgaP | | 著者 | Eichmann, C, Tzitzilonis, C, Nakamura, T, Kwiatkowski, W, Maslennikov, I, Choe, S, Lipton, S.A, Riek, R. | | 登録日 | 2016-01-01 | | 公開日 | 2016-08-10 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
5LAO
 
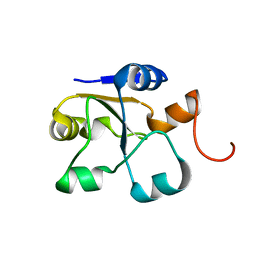 | | S-nitrosylated 3D NMR structure of the cytoplasmic rhodanese domain of the inner membrane protein YgaP from Escherichia coli | | 分子名称: | Inner membrane protein YgaP | | 著者 | Eichmann, C, Tzitzilonis, C, Nakamura, T, Maslennikov, I, Kwiatkowski, W, Choe, S, Lipton, S.A, Guntert, P, Riek, R. | | 登録日 | 2016-06-14 | | 公開日 | 2016-08-17 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
5LAM
 
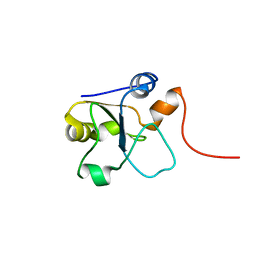 | | Refined 3D NMR structure of the cytoplasmic rhodanese domain of the inner membrane protein YgaP from Escherichia coli | | 分子名称: | Inner membrane protein YgaP | | 著者 | Eichmann, C, Tzitzilonis, C, Nakamura, T, Maslennikov, I, Kwiatkowski, W, Choe, S, Lipton, S.A, Guntert, P, Riek, R. | | 登録日 | 2016-06-14 | | 公開日 | 2016-08-17 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
