1Z66
 
 | | NMR solution structure of domain III of E-protein of tick-borne Langat flavivirus (no RDC restraints) | | 分子名称: | Major envelope protein E | | 著者 | Mukherjee, M, Dutta, K, White, M.A, Cowburn, D, Fox, R.O. | | 登録日 | 2005-03-21 | | 公開日 | 2006-03-28 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR solution structure and backbone dynamics of domain III of the E protein of tick-borne Langat flavivirus suggests a potential site for molecular recognition.
Protein Sci., 15, 2006
|
|
1QZM
 
 | | alpha-domain of ATPase | | 分子名称: | ATP-dependent protease La | | 著者 | Botos, I, Melnikov, E.E, Cherry, S, Khalatova, A.G, Rasulova, F.S, Tropea, J.E, Maurizi, M.R, Rotanova, T.V, Gustchina, A, Wlodawer, A. | | 登録日 | 2003-09-17 | | 公開日 | 2004-05-04 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of the AAA+ alpha domain of E. coli Lon protease at 1.9A resolution.
J.Struct.Biol., 146
|
|
5FKA
 
 | | Crystal structure of staphylococcal enterotoxin E in complex with a T cell receptor | | 分子名称: | STAPHYLOCOCCAL ENTEROTOXIN E, T CELL RECEPTOR ALPHA CHAIN, T CELL RECEPTOR BETA CHAIN, ... | | 著者 | Rodstrom, K.E.J, Regenthal, P, Lindkvist-Petersson, K. | | 登録日 | 2015-10-15 | | 公開日 | 2016-05-25 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Two Common Structural Motifs for Tcr Recognition by Staphylococcal Enterotoxins.
Sci.Rep., 6, 2016
|
|
5G1C
 
 | | Structure of HDAC like protein from Bordetella Alcaligenes bound the photoswitchable pyrazole Inhibitor CEW395 | | 分子名称: | (2E)-N-hydroxy-3-{4-[(E)-(1,3,5-trimethyl-1H-pyrazol-4-yl)diazenyl]phenyl}prop-2-enamide, DI(HYDROXYETHYL)ETHER, HISTONE DEACETYLASE-LIKE AMIDOHYDROLASE, ... | | 著者 | Kraemer, A, Meyer-Almes, F.J, Yildiz, O. | | 登録日 | 2016-03-24 | | 公開日 | 2016-11-23 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Toward Photopharmacological Antimicrobial Chemotherapy Using Photoswitchable Amidohydrolase Inhibitors.
ACS Infect Dis, 3, 2017
|
|
5N2R
 
 | | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with PSB36 at 2.8A resolution | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-butyl-3-[(~{E})-3-oxidanylprop-1-enyl]-8-[(1~{R},5~{S})-3-tricyclo[3.3.1.0^{3,7}]nonanyl]-7~{H}-purine-2,6-dione, Adenosine receptor A2a,Soluble cytochrome b562,ADENOSINE RECEPTOR A2A SOLUBLE CYTOCHROME B562 ADENOSINE RECEPTOR A2A,Adenosine receptor A2a, ... | | 著者 | Cheng, R.K.Y, Segala, E, Robertson, N, Deflorian, F, Dore, A.S, Errey, J.C, Fiez-Vandal, C, Marshall, F.H, Cooke, R.M. | | 登録日 | 2017-02-08 | | 公開日 | 2017-07-26 | | 最終更新日 | 2017-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structures of Human A1 and A2A Adenosine Receptors with Xanthines Reveal Determinants of Selectivity.
Structure, 25, 2017
|
|
7W71
 
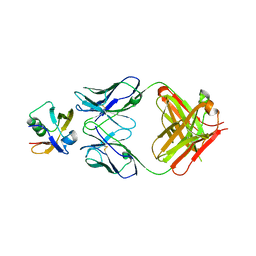 | |
3L6Y
 
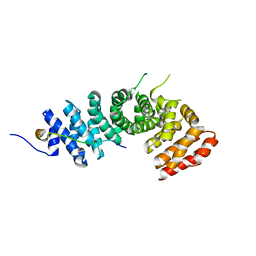 | | Crystal structure of p120 catenin in complex with E-cadherin | | 分子名称: | Catenin delta-1, E-cadherin | | 著者 | Ishiyama, N, Lee, S.-H, Liu, S, Li, G.-Y, Smith, M.J, Reichardt, L.F, Ikura, M. | | 登録日 | 2009-12-27 | | 公開日 | 2010-04-21 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion.
Cell(Cambridge,Mass.), 141, 2010
|
|
4YJ2
 
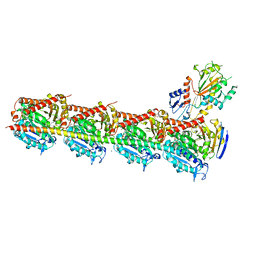 | | Crystal structure of tubulin bound to MI-181 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5,6-dimethyl-2-[(E)-2-(pyridin-3-yl)ethenyl]-1,3-benzothiazole, CALCIUM ION, ... | | 著者 | McNamara, D.E, Torres, J.Z, Yeates, T.O. | | 登録日 | 2015-03-03 | | 公開日 | 2015-05-27 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structures of potent anticancer compounds bound to tubulin.
Protein Sci., 24, 2015
|
|
3L6X
 
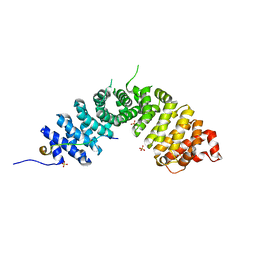 | | Crystal structure of p120 catenin in complex with E-cadherin | | 分子名称: | Catenin delta-1, E-cadherin, SULFATE ION | | 著者 | Ishiyama, N, Lee, S.-H, Liu, S, Li, G.-Y, Smith, M.J, Reichardt, L.F, Ikura, M. | | 登録日 | 2009-12-27 | | 公開日 | 2010-04-21 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion.
Cell(Cambridge,Mass.), 141, 2010
|
|
5G3W
 
 | | Structure of HDAC like protein from Bordetella Alcaligenes in complex with the photoswitchable inhibitor CEW65 | | 分子名称: | (2E)-N-hydroxy-3-{4-[(E)-phenyldiazenyl]phenyl}prop-2-enamide, DI(HYDROXYETHYL)ETHER, HISTONE DEACETYLASE-LIKE AMIDOHYDROLASE, ... | | 著者 | Kraemer, A, Meyer-Almes, F.J, Yildiz, O. | | 登録日 | 2016-05-02 | | 公開日 | 2016-11-23 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Toward Photopharmacological Antimicrobial Chemotherapy Using Photoswitchable Amidohydrolase Inhibitors.
ACS Infect Dis, 3, 2017
|
|
6QE5
 
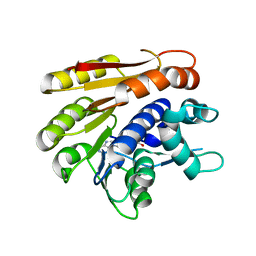 | | Structure of E.coli RlmJ in complex with the natural cofactor product S-adenosyl-homocysteine (SAH) | | 分子名称: | Ribosomal RNA large subunit methyltransferase J, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Oerum, S, Catala, M, Atdjian, C, Brachet, F, Ponchon, L, Barraud, P, Iannazzo, L, Droogmans, L, Braud, E, Etheve-Quelquejeu, M, Tisne, C. | | 登録日 | 2019-01-04 | | 公開日 | 2019-03-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Bisubstrate analogues as structural tools to investigate m6A methyltransferase active sites.
Rna Biol., 16, 2019
|
|
6QDX
 
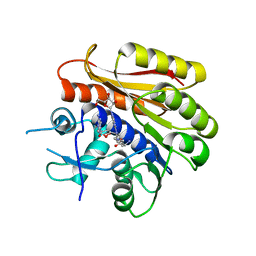 | | Structure of E.coli RlmJ in complex with a bisubstrate analogue (BA4) | | 分子名称: | (2~{S})-4-[[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-[3-[[9-[(2~{S},3~{R},4~{S},5~{S})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]purin-6-yl]amino]propyl]amino]-2-azanyl-butanoic acid, Ribosomal RNA large subunit methyltransferase J | | 著者 | Oerum, S, Catala, M, Atdjian, C, Brachet, F, Ponchon, L, Barraud, P, Iannazzo, L, Droogmans, L, Braud, E, Etheve-Quelquejeu, M, Tisne, C. | | 登録日 | 2019-01-03 | | 公開日 | 2019-03-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Bisubstrate analogues as structural tools to investigate m6A methyltransferase active sites.
Rna Biol., 16, 2019
|
|
5ZDP
 
 | | Crystal structure of cyanide-insensitive alternative oxidase from Trypanosoma brucei with ferulenol | | 分子名称: | 4-oxidanyl-3-[(2~{E},6~{E})-3,7,11-trimethyldodeca-2,6,10-trienyl]chromen-2-one, Alternative oxidase, mitochondrial, ... | | 著者 | Shiba, T, Inaoka, D.K, Takahashi, G, Tsuge, C, Kido, Y, Young, L, Ueda, S, Balogun, E.O, Nara, T, Honma, T, Tanaka, A, Inoue, M, Saimoto, H, Harada, S, Moore, A.L, Kita, K. | | 登録日 | 2018-02-23 | | 公開日 | 2019-02-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Insights into the ubiquinol/dioxygen binding and proton relay pathways of the alternative oxidase.
Biochim Biophys Acta Bioenerg, 1860, 2019
|
|
6L0R
 
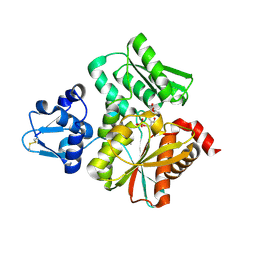 | | Crystal Structure of the O-Phosphoserine Sulfhydrylase from Aeropyrum pernix Complexed with O-Acetylserine | | 分子名称: | (2S)-3-acetyloxy-2-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]propanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Protein CysO | | 著者 | Nakabayashi, M, Takeda, E, Ishikawa, K, Nakamura, T. | | 登録日 | 2019-09-26 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Identification of amino acid residues important for recognition of O-phospho-l-serine substrates by cysteine synthase.
J.Biosci.Bioeng., 131, 2021
|
|
3V81
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) with DNA and the nonnucleoside inhibitor nevirapine | | 分子名称: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, DNA (5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(ATM))-3'), DNA (5'-D(*AP*TP*GP*GP*AP*AP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), ... | | 著者 | Das, K, Martinez, S.E, Arnold, E. | | 登録日 | 2011-12-22 | | 公開日 | 2012-01-18 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.8503 Å) | | 主引用文献 | HIV-1 reverse transcriptase complex with DNA and nevirapine reveals non-nucleoside inhibition mechanism.
Nat.Struct.Mol.Biol., 19, 2012
|
|
6A6K
 
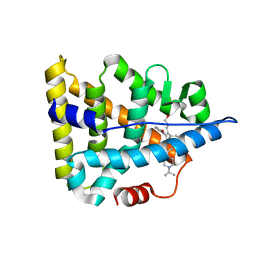 | | Crystal structure of Estrogen-related Receptor-3 (ERR-gamma) ligand binding domain with DN201000 | | 分子名称: | 3-[(~{E})-5-oxidanyl-2-phenyl-1-[4-(4-propan-2-ylpiperazin-1-yl)phenyl]pent-1-enyl]phenol, Estrogen-related receptor gamma | | 著者 | Yoon, H, Kim, J, Chin, J, Cho, S.J, Song, J. | | 登録日 | 2018-06-28 | | 公開日 | 2019-04-10 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Discovery of Potent, Selective, and Orally Bioavailable Estrogen-Related Receptor-gamma Inverse Agonists To Restore the Sodium Iodide Symporter Function in Anaplastic Thyroid Cancer.
J. Med. Chem., 62, 2019
|
|
6AD9
 
 | | Crystal Structure of PPARgamma Ligand Binding Domain in complex with dibenzooxepine derivative compound-9 | | 分子名称: | 12-mer peptide from Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, 3-[(1E)-1-{8-[(4-methyl-2-propyl-1H-benzimidazol-1-yl)methyl]dibenzo[b,e]oxepin-11(6H)-ylidene}ethyl]-1,2,4-oxadiazol-5(4H)-one, Peroxisome proliferator-activated receptor gamma | | 著者 | Takahashi, Y, Suzuki, M, Yamamoto, K, Saito, J. | | 登録日 | 2018-07-31 | | 公開日 | 2018-11-14 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Development of Dihydrodibenzooxepine Peroxisome Proliferator-Activated Receptor (PPAR) Gamma Ligands of a Novel Binding Mode as Anticancer Agents: Effective Mimicry of Chiral Structures by Olefinic E/ Z-Isomers.
J. Med. Chem., 61, 2018
|
|
6L0S
 
 | | Crystal Structure of the O-Phosphoserine Sulfhydrylase from Aeropyrum pernix Complexed with L-Cysteine | | 分子名称: | (2R)-2-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-3-sulfanyl-propanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Protein CysO | | 著者 | Nakabayashi, M, Takeda, E, Ishikawa, K, Nakamura, T. | | 登録日 | 2019-09-26 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Identification of amino acid residues important for recognition of O-phospho-l-serine substrates by cysteine synthase.
J.Biosci.Bioeng., 131, 2021
|
|
6L0Q
 
 | | Crystal Structure of the O-Phosphoserine Sulfhydrylase from Aeropyrum pernix Complexed with O-Phosphoserine | | 分子名称: | (2S)-2-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-3-phosphonooxy-propanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Protein CysO | | 著者 | Nakabayashi, M, Takeda, E, Ishikawa, K, Nakamura, T. | | 登録日 | 2019-09-26 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Identification of amino acid residues important for recognition of O-phospho-l-serine substrates by cysteine synthase.
J.Biosci.Bioeng., 131, 2021
|
|
6L0P
 
 | | Crystal Structure of the O-Phosphoserine Sulfhydrylase from Aeropyrum pernix Complexed with O-Phosphoserine | | 分子名称: | (2S)-2-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-3-phosphonooxy-propanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Protein CysO | | 著者 | Nakabayashi, M, Takeda, E, Ishikawa, K, Nakamura, T. | | 登録日 | 2019-09-26 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Identification of amino acid residues important for recognition of O-phospho-l-serine substrates by cysteine synthase.
J.Biosci.Bioeng., 131, 2021
|
|
2XO5
 
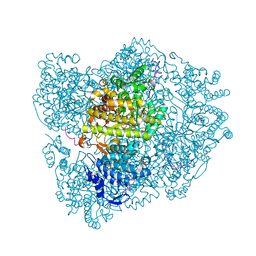 | | RIBONUCLEOTIDE REDUCTASE Y731NH2Y MODIFIED R1 SUBUNIT OF E. COLI | | 分子名称: | RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 SUBUNIT ALPHA, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 SUBUNIT BETA | | 著者 | Minnihan, E.C, Seyedsayamdost, M.R, Uhlin, U, Stubbe, J. | | 登録日 | 2010-08-09 | | 公開日 | 2010-08-18 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Kinetics of Radical Intermediate Formation and Deoxynucleotide Production in 3-Aminotyrosine- Substituted Escherichia Coli Ribonucleotide Reductases.
J.Am.Chem.Soc., 133, 2011
|
|
6CWD
 
 | |
7N6K
 
 | | Crystal structure of the substrate-binding domain of E. coli DnaK in complex with the peptide RALALLPLSR | | 分子名称: | Alkaline phosphatase peptide, Chaperone protein DnaK, SULFATE ION | | 著者 | Jansen, R.M, Ozden, C, Gierasch, L.M, Garman, S.C. | | 登録日 | 2021-06-08 | | 公開日 | 2021-06-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Selective promiscuity in the binding of E. coli Hsp70 to an unfolded protein.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N6J
 
 | | Crystal structure of the substrate-binding domain of E. coli DnaK in complex with the peptide RKQSTIALALLPLLFTPRR | | 分子名称: | Alkaline phosphatase peptide, Chaperone protein DnaK, SULFATE ION | | 著者 | Jansen, R.M, Ozden, C, Gierasch, L.M, Garman, S.C. | | 登録日 | 2021-06-08 | | 公開日 | 2021-06-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Selective promiscuity in the binding of E. coli Hsp70 to an unfolded protein.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5FKZ
 
 | | Structure of E.coli Constitutive lysine decarboxylase | | 分子名称: | LYSINE DECARBOXYLASE, CONSTITUTIVE | | 著者 | Kandiah, E, Carriel, D, Perard, J, Malet, H, Bacia, M, Liu, K, Chan, S.W.S, Houry, W.A, Ollagnier de Choudens, S, Elsen, S, Gutsche, I. | | 登録日 | 2015-10-20 | | 公開日 | 2016-09-21 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (5.5 Å) | | 主引用文献 | Structural Insights Into the Escherichia Coli Lysine Decarboxylases and Molecular Determinants of Interaction with the Aaa+ ATPase Rava.
Sci.Rep., 6, 2016
|
|
