1BQM
 
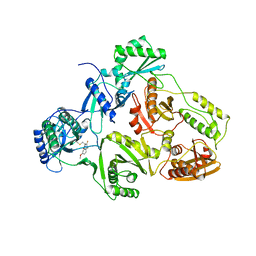 | | HIV-1 RT/HBY 097 | | 分子名称: | (S)-4-ISOPROPOXYCARBONYL-6-METHOXY-3-METHYLTHIOMETHYL-3,4-DIHYDROQUINOXALIN-2(1H)-THIONE, REVERSE TRANSCRIPTASE | | 著者 | Hsiou, Y, Das, K, Ding, J, Arnold, E. | | 登録日 | 1998-08-17 | | 公開日 | 1999-01-06 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structures of Tyr188Leu mutant and wild-type HIV-1 reverse transcriptase complexed with the non-nucleoside inhibitor HBY 097: inhibitor flexibility is a useful design feature for reducing drug resistance.
J.Mol.Biol., 284, 1998
|
|
1BQN
 
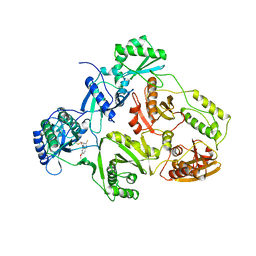 | | TYR 188 LEU HIV-1 RT/HBY 097 | | 分子名称: | (S)-4-ISOPROPOXYCARBONYL-6-METHOXY-3-METHYLTHIOMETHYL-3,4-DIHYDROQUINOXALIN-2(1H)-THIONE, REVERSE TRANSCRIPTASE | | 著者 | Hsiou, Y, Das, K, Ding, J, Arnold, E. | | 登録日 | 1998-08-17 | | 公開日 | 1999-01-06 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structures of Tyr188Leu mutant and wild-type HIV-1 reverse transcriptase complexed with the non-nucleoside inhibitor HBY 097: inhibitor flexibility is a useful design feature for reducing drug resistance.
J.Mol.Biol., 284, 1998
|
|
1BQO
 
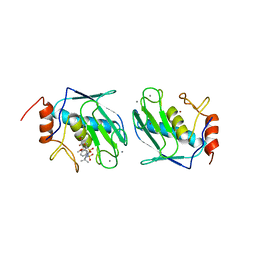 | | DISCOVERY OF POTENT, ACHIRAL MATRIX METALLOPROTEINASE INHIBITORS | | 分子名称: | 1,3-BIS-(4-METHOXY-BENZENESULFONYL)-5,5-DIMETHYL-HEXAHYDRO-PYRIMIDINE-2-CARBOXYLIC ACID HYDROXYAMIDE, CALCIUM ION, STROMELYSIN-1, ... | | 著者 | Pikul, S, Dunham, K.L.M, Almstead, N.G, De, B, Natchus, M.G, Anastasio, M.V, Mcphail, S.J, Snider, C.E, Taiwo, Y.O, Rydel, T.J, Dunaway, C.M, Gu, F, Mieling, G.E. | | 登録日 | 1998-08-17 | | 公開日 | 1999-08-17 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Discovery of potent, achiral matrix metalloproteinase inhibitors.
J.Med.Chem., 41, 1998
|
|
1GFS
 
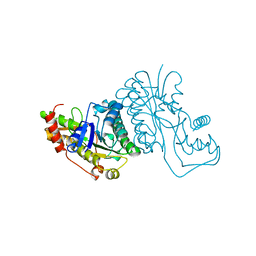 | | GDP-FUCOSE SYNTHETASE FROM E. COLI | | 分子名称: | GDP-FUCOSE SYNTHETASE | | 著者 | Somers, W.S, Stahl, M.L, Sullivan, F.X. | | 登録日 | 1998-08-17 | | 公開日 | 1999-08-17 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | GDP-fucose synthetase from Escherichia coli: structure of a unique member of the short-chain dehydrogenase/reductase family that catalyzes two distinct reactions at the same active site.
Structure, 6, 1998
|
|
3GRX
 
 | | NMR STRUCTURE OF ESCHERICHIA COLI GLUTAREDOXIN 3-GLUTATHIONE MIXED DISULFIDE COMPLEX, 20 STRUCTURES | | 分子名称: | GLUTAREDOXIN 3, GLUTATHIONE | | 著者 | Nordstrand, K, Aslund, F, Holmgren, A, Otting, G, Berndt, K.D. | | 登録日 | 1998-08-17 | | 公開日 | 1999-03-30 | | 最終更新日 | 2018-03-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of Escherichia coli glutaredoxin 3-glutathione mixed disulfide complex: implications for the enzymatic mechanism.
J.Mol.Biol., 286, 1999
|
|
2MYO
 
 | | SOLUTION STRUCTURE OF MYOTROPHIN, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | MYOTROPHIN | | 著者 | Yang, Y, Nanduri, S, Sen, S, Qin, J. | | 登録日 | 1998-08-17 | | 公開日 | 1999-08-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The structural basis of ankyrin-like repeat function as revealed by the solution structure of myotrophin.
Structure, 6, 1998
|
|
1MHD
 
 | |
1BQU
 
 | | CYTOKYNE-BINDING REGION OF GP130 | | 分子名称: | GLYCEROL, PROTEIN (GP130), SULFATE ION | | 著者 | Bravo, J, Staunton, D, Heath, J.K, Jones, E.Y. | | 登録日 | 1998-08-18 | | 公開日 | 1998-08-26 | | 最終更新日 | 2022-12-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of a cytokine-binding region of gp130.
EMBO J., 17, 1998
|
|
1BQS
 
 | | THE CRYSTAL STRUCTURE OF MUCOSAL ADDRESSIN CELL ADHESION MOLECULE-1 (MADCAM-1) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (MUCOSAL ADDRESSIN CELL ADHESION MOLECULE-1) | | 著者 | Tan, K, Casasnovas, J.M, Liu, J.H, Briskin, M.J, Springer, T.A, Wang, J.-H. | | 登録日 | 1998-08-18 | | 公開日 | 1999-08-13 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The structure of immunoglobulin superfamily domains 1 and 2 of MAdCAM-1 reveals novel features important for integrin recognition.
Structure, 6, 1998
|
|
1BQT
 
 | | THREE-DIMENSIONAL STRUCTURE OF HUMAN INSULIN-LIKE GROWTH FACTOR-I (IGF-I) DETERMINED BY 1H-NMR AND DISTANCE GEOMETRY, 6 STRUCTURES | | 分子名称: | INSULIN-LIKE GROWTH FACTOR-I | | 著者 | Sato, A, Nishimura, S, Ohkubo, T, Kyogoku, Y, Koyama, S, Kobayashi, M, Yasuda, T, Kobayashi, Y. | | 登録日 | 1998-08-18 | | 公開日 | 1999-05-18 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-dimensional structure of human insulin-like growth factor-I (IGF-I) determined by 1H-NMR and distance geometry.
Int.J.Pept.Protein Res., 41, 1993
|
|
1BQQ
 
 | | CRYSTAL STRUCTURE OF THE MT1-MMP--TIMP-2 COMPLEX | | 分子名称: | CALCIUM ION, MEMBRANE-TYPE MATRIX METALLOPROTEINASE, METALLOPROTEINASE INHIBITOR 2, ... | | 著者 | Fernandez-Catalan, C, Bode, W, Huber, R, Turk, D, Calvete, J.J, Lichte, A, Tschesche, H, Maskos, K. | | 登録日 | 1998-08-18 | | 公開日 | 1999-08-18 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Crystal structure of the complex formed by the membrane type 1-matrix metalloproteinase with the tissue inhibitor of metalloproteinases-2, the soluble progelatinase A receptor.
EMBO J., 17, 1998
|
|
1LDZ
 
 | |
2LDZ
 
 | |
5PGM
 
 | |
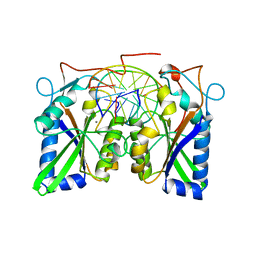
3BAM
 
 | |
2ARG
 
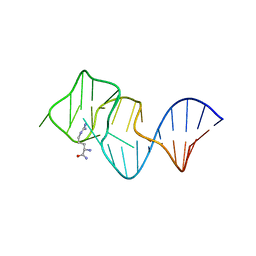 | | FORMATION OF AN AMINO ACID BINDING POCKET THROUGH ADAPTIVE ZIPPERING-UP OF A LARGE DNA HAIRPIN LOOP, NMR, 9 STRUCTURES | | 分子名称: | ARGININEAMIDE, DNA APTAMER [5'-D (*TP*GP*AP*CP*CP*AP*GP*GP*GP*CP*AP*AP*AP*CP*GP*GP*TP*AP* GP*GP*TP*GP*AP*GP*TP*GP*GP*TP*CP*A)-3'] | | 著者 | Lin, C.H, Wang, W, Jones, R.A, Patel, D.J. | | 登録日 | 1998-08-19 | | 公開日 | 1999-03-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Formation of an amino-acid-binding pocket through adaptive zippering-up of a large DNA hairpin loop.
Chem.Biol., 5, 1998
|
|
2BAM
 
 | |
1SKF
 
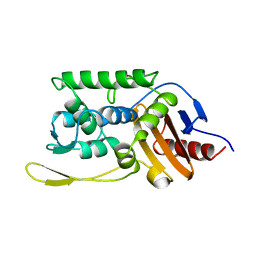 | |
1MUD
 
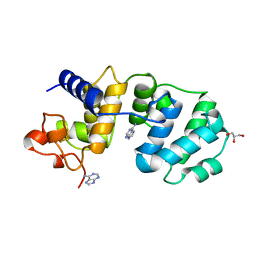 | | CATALYTIC DOMAIN OF MUTY FROM ESCHERICHIA COLI, D138N MUTANT COMPLEXED TO ADENINE | | 分子名称: | ADENINE, Adenine DNA glycosylase, GLYCEROL, ... | | 著者 | Guan, Y, Tainer, J.A. | | 登録日 | 1998-08-20 | | 公開日 | 1999-09-20 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily.
Nat.Struct.Biol., 5, 1998
|
|
1NK1
 
 | | NK1 FRAGMENT OF HUMAN HEPATOCYTE GROWTH FACTOR/SCATTER FACTOR (HGF/SF) AT 2.5 ANGSTROM RESOLUTION | | 分子名称: | PROTEIN (HEPATOCYTE GROWTH FACTOR PRECURSOR) | | 著者 | Chirgadze, D.Y, Hepple, J.P, Zhou, H, Byrd, R.A, Blundell, T.L, Gherardi, E. | | 登録日 | 1998-08-20 | | 公開日 | 1999-01-13 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of the NK1 fragment of HGF/SF suggests a novel mode for growth factor dimerization and receptor binding.
Nat.Struct.Biol., 6, 1999
|
|
1MUY
 
 | | CATALYTIC DOMAIN OF MUTY FROM ESCHERICHIA COLI | | 分子名称: | ADENINE GLYCOSYLASE, GLYCEROL, IMIDAZOLE, ... | | 著者 | Guan, Y, Tainer, J.A. | | 登録日 | 1998-08-20 | | 公開日 | 1999-08-20 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily.
Nat.Struct.Biol., 5, 1998
|
|
5PAH
 
 | |
1PBI
 
 | |
6PAH
 
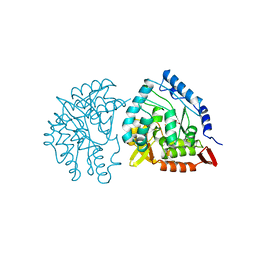 | | HUMAN PHENYLALANINE HYDROXYLASE CATALYTIC DOMAIN DIMER WITH BOUND L-DOPA (3,4-DIHYDROXYPHENYLALANINE) INHIBITOR | | 分子名称: | 3,4-DIHYDROXYPHENYLALANINE, FE (III) ION, PHENYLALANINE 4-MONOOXYGENASE | | 著者 | Erlandsen, H, Flatmark, T, Stevens, R.C. | | 登録日 | 1998-08-20 | | 公開日 | 1999-04-27 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystallographic analysis of the human phenylalanine hydroxylase catalytic domain with bound catechol inhibitors at 2.0 A resolution.
Biochemistry, 37, 1998
|
|
1BQ3
 
 | |
