7TDD
 
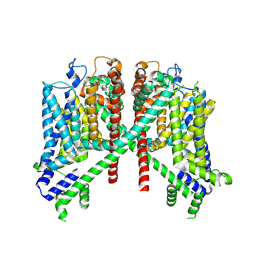 | | AtTPC1 D454N-EDTA state II | | 分子名称: | Two pore calcium channel protein 1 | | 著者 | Dickinson, M.S, Stroud, R.M. | | 登録日 | 2021-12-30 | | 公開日 | 2022-10-12 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Molecular basis of multistep voltage activation in plant two-pore channel 1.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7TDE
 
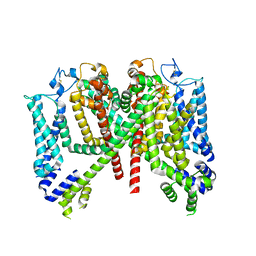 | |
2AYA
 
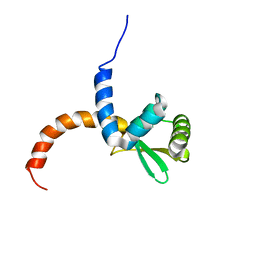 | |
8Q1M
 
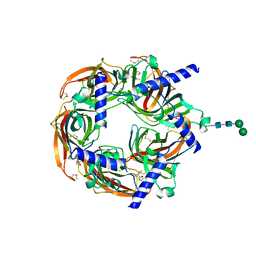 | | Aplysia californica acetylcholine-binding protein in complex with Spiroimine (+)-4 R | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Soluble acetylcholine receptor, ... | | 著者 | Sulzenbacher, G, Bourne, Y, Marchot, P. | | 登録日 | 2023-07-31 | | 公開日 | 2024-04-03 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The Cyclic Imine Core Common to the Marine Macrocyclic Toxins Is Sufficient to Dictate Nicotinic Acetylcholine Receptor Antagonism.
Mar Drugs, 22, 2024
|
|
2AXK
 
 | | Solution structure of discrepin, a scorpion venom toxin blocking K+ channels. | | 分子名称: | discrepin | | 著者 | Prochnicka-Chalufour, A, Corzo, G, Satake, H, Martin-Eauclaire, M.-F, Murgia, A.R, Prestipino, G, D'Suze, G, Possani, L.D, Delepierre, M. | | 登録日 | 2005-09-05 | | 公開日 | 2006-06-20 | | 最終更新日 | 2019-12-25 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of discrepin, a new K+-channel blocking peptide from the alpha-KTx15 subfamily.
Biochemistry, 45, 2006
|
|
2AL4
 
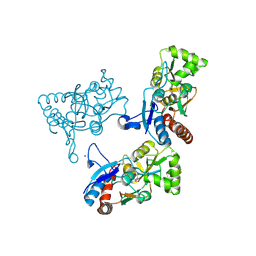 | | CRYSTAL STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH quisqualate and CX614. | | 分子名称: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 2,3,6A,7,8,9-HEXAHYDRO-11H-[1,4]DIOXINO[2,3-G]PYRROLO[2,1-B][1,3]BENZOXAZIN-11-ONE, Glutamate receptor 2, ... | | 著者 | Jin, R, Clark, S, Weeks, A.M, Dudman, J.T, Gouaux, E, Partin, K.M. | | 登録日 | 2005-08-04 | | 公開日 | 2005-10-25 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Mechanism of positive allosteric modulators acting on AMPA receptors.
J.Neurosci., 25, 2005
|
|
2CWW
 
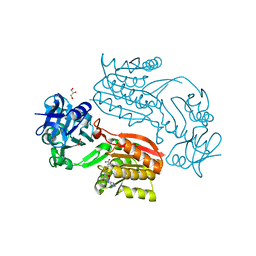 | | Crystal structure of Thermus thermophilus TTHA1280, a putative SAM-dependent RNA methyltransferase, in complex with S-adenosyl-L-homocysteine | | 分子名称: | ACETIC ACID, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | 著者 | Pioszak, A.A, Murayama, K, Nakagawa, N, Ebihara, A, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2005-06-27 | | 公開日 | 2005-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structures of a putative RNA 5-methyluridine methyltransferase, Thermus thermophilus TTHA1280, and its complex with S-adenosyl-L-homocysteine.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
4DAM
 
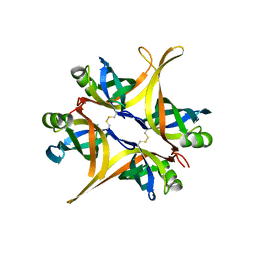 | | Crystal structure of small single-stranded DNA-binding protein from Streptomyces coelicolor | | 分子名称: | Single-stranded DNA-binding protein 1 | | 著者 | Filic, Z, Herron, P, Ivic, N, Luic, M, Manjasetty, B.A, Paradzik, T, Vujaklija, D. | | 登録日 | 2012-01-13 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure-function relationships of two paralogous single-stranded DNA-binding proteins from Streptomyces coelicolor: implication of SsbB in chromosome segregation during sporulation.
Nucleic Acids Res., 41, 2013
|
|
3H6W
 
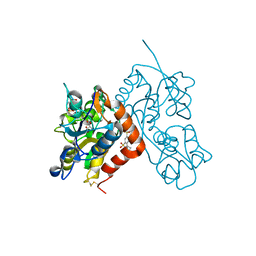 | | Crystal structure of the iGluR2 ligand-binding core (S1S2J-N754S) in complex with glutamate and NS5217 at 1.50 A resolution | | 分子名称: | (3R)-3-cyclopentyl-6-methyl-7-[(4-methylpiperazin-1-yl)sulfonyl]-3,4-dihydro-2H-1,2-benzothiazine 1,1-dioxide, DIMETHYL SULFOXIDE, GLUTAMIC ACID, ... | | 著者 | Hald, H, Gajhede, M, Kastrup, J.S. | | 登録日 | 2009-04-24 | | 公開日 | 2009-07-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Distinct structural features of cyclothiazide are responsible for effects on peak current amplitude and desensitization kinetics at iGluR2.
J.Mol.Biol., 391, 2009
|
|
7C17
 
 | |
3J8B
 
 | |
3J8C
 
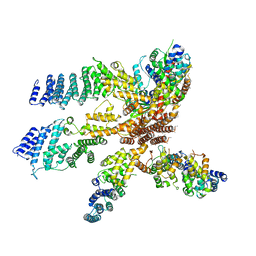 | | Model of the human eIF3 PCI-MPN octamer docked into the 43S EM map | | 分子名称: | Eukaryotic translation initiation factor 3 subunit A, Eukaryotic translation initiation factor 3 subunit C, Eukaryotic translation initiation factor 3 subunit E, ... | | 著者 | Erzberger, J.P, Ban, N. | | 登録日 | 2014-10-08 | | 公開日 | 2014-10-22 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (11.6 Å) | | 主引用文献 | Molecular Architecture of the 40SeIF1eIF3 Translation Initiation Complex.
Cell(Cambridge,Mass.), 158, 2014
|
|
3H6T
 
 | | Crystal structure of the iGluR2 ligand-binding core (S1S2J-N754S) in complex with glutamate and cyclothiazide at 2.25 A resolution | | 分子名称: | ACETATE ION, CACODYLATE ION, CYCLOTHIAZIDE, ... | | 著者 | Hald, H, Gajhede, M, Kastrup, J.S. | | 登録日 | 2009-04-24 | | 公開日 | 2009-07-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Distinct structural features of cyclothiazide are responsible for effects on peak current amplitude and desensitization kinetics at iGluR2.
J.Mol.Biol., 391, 2009
|
|
3H6V
 
 | | Crystal structure of the iGluR2 ligand-binding core (S1S2J-N754S) in complex with glutamate and NS5206 at 2.10 A resolution | | 分子名称: | (3R)-3-cyclopentyl-7-[(4-methylpiperazin-1-yl)sulfonyl]-3,4-dihydro-2H-1,2-benzothiazine 1,1-dioxide, DIMETHYL SULFOXIDE, GLUTAMIC ACID, ... | | 著者 | Hald, H, Gajhede, M, Kastrup, J.S. | | 登録日 | 2009-04-24 | | 公開日 | 2009-07-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Distinct structural features of cyclothiazide are responsible for effects on peak current amplitude and desensitization kinetics at iGluR2.
J.Mol.Biol., 391, 2009
|
|
3H6U
 
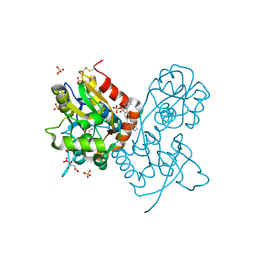 | | Crystal structure of the iGluR2 ligand-binding core (S1S2J-N754S) in complex with glutamate and NS1493 at 1.85 A resolution | | 分子名称: | (3S)-3-cyclopentyl-6-methyl-7-[(4-methylpiperazin-1-yl)sulfonyl]-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, CITRATE ANION, GLUTAMIC ACID, ... | | 著者 | Hald, H, Gajhede, M, Kastrup, J.S. | | 登録日 | 2009-04-24 | | 公開日 | 2009-07-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Distinct structural features of cyclothiazide are responsible for effects on peak current amplitude and desensitization kinetics at iGluR2.
J.Mol.Biol., 391, 2009
|
|
6XJF
 
 | |
7OZN
 
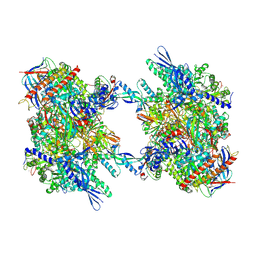 | | RNA Polymerase II dimer (Class 1) | | 分子名称: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | 著者 | Aibara, S, Dienemann, C, Cramer, P. | | 登録日 | 2021-06-28 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structure of an inactive RNA polymerase II dimer.
Nucleic Acids Res., 49, 2021
|
|
7OZP
 
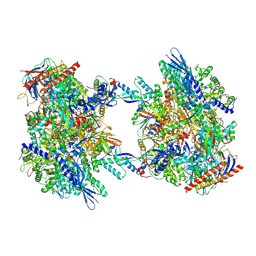 | | RNA Polymerase II dimer (Class 3) | | 分子名称: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | 著者 | Aibara, S, Dienemann, C, Cramer, P. | | 登録日 | 2021-06-28 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure of an inactive RNA polymerase II dimer.
Nucleic Acids Res., 49, 2021
|
|
7OOP
 
 | | Pol II-CSB-CSA-DDB1-UVSSA-PAF-SPT6 (Structure 3) | | 分子名称: | DNA damage-binding protein 1, DNA excision repair protein ERCC-6, DNA excision repair protein ERCC-8, ... | | 著者 | Kokic, G, Cramer, P. | | 登録日 | 2021-05-28 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis of human transcription-DNA repair coupling.
Nature, 598, 2021
|
|
6XRE
 
 | | Structure of the p53/RNA polymerase II assembly | | 分子名称: | Cellular tumor antigen p53, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11-a, ... | | 著者 | Liou, S.-H, Singh, S, Singer, R.H, Coleman, R.A, Liu, W. | | 登録日 | 2020-07-12 | | 公開日 | 2021-03-24 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Structure of the p53/RNA polymerase II assembly.
Commun Biol, 4, 2021
|
|
7OZO
 
 | | RNA Polymerase II dimer (Class 2) | | 分子名称: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | 著者 | Aibara, S, Dienemann, C, Cramer, P. | | 登録日 | 2021-06-28 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure of an inactive RNA polymerase II dimer.
Nucleic Acids Res., 49, 2021
|
|
7OO3
 
 | | Pol II-CSB-CSA-DDB1-UVSSA (Structure1) | | 分子名称: | CSB element, DNA damage-binding protein 1, DNA excision repair protein ERCC-6, ... | | 著者 | Kokic, G, Cramer, P. | | 登録日 | 2021-05-26 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural basis of human transcription-DNA repair coupling.
Nature, 598, 2021
|
|
7OPD
 
 | |
7OOB
 
 | |
7OPC
 
 | |
