1F5Q
 
 | | CRYSTAL STRUCTURE OF MURINE GAMMA HERPESVIRUS CYCLIN COMPLEXED TO HUMAN CYCLIN DEPENDENT KINASE 2 | | 分子名称: | CHLORIDE ION, CYCLIN DEPENDENT KINASE 2, GAMMA HERPESVIRUS CYCLIN | | 著者 | Card, G.L, Knowles, P, Laman, H, Jones, N, McDonald, N.Q. | | 登録日 | 2000-06-15 | | 公開日 | 2000-12-27 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of a gamma-herpesvirus cyclin-cdk complex.
EMBO J., 19, 2000
|
|
1G5S
 
 | | CRYSTAL STRUCTURE OF HUMAN CYCLIN DEPENDENT KINASE 2 (CDK2) IN COMPLEX WITH THE INHIBITOR H717 | | 分子名称: | 2-[TRANS-(4-AMINOCYCLOHEXYL)AMINO]-6-(BENZYL-AMINO)-9-CYCLOPENTYLPURINE, CELL DIVISION PROTEIN KINASE 2 | | 著者 | Dreyer, M.K, Borcherding, D.R, Dumont, J.A, Peet, N.P, Tsay, J.T, Wright, P.S, Bitonti, A.J, Shen, J, Kim, S.-H. | | 登録日 | 2000-11-02 | | 公開日 | 2001-11-02 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Crystal structure of human cyclin-dependent kinase 2 in complex with the adenine-derived inhibitor H717.
J.Med.Chem., 44, 2001
|
|
1GIJ
 
 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE CDK4 INHIBITOR | | 分子名称: | 1-(5-OXO-2,3,5,9B-TETRAHYDRO-1H-PYRROLO[2,1-A]ISOINDOL-9-YL)-3-(5-PYRROLIDIN-2-YL-1H-PYRAZOL-3-YL)-UREA, CELL DIVISION PROTEIN KINASE 2 | | 著者 | Ikuta, M, Nishimura, S. | | 登録日 | 2001-02-06 | | 公開日 | 2002-02-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystallographic approach to identification of cyclin-dependent kinase 4 (CDK4)-specific inhibitors by using CDK4 mimic CDK2 protein.
J.Biol.Chem., 276, 2002
|
|
1GH6
 
 | |
1FVV
 
 | | THE STRUCTURE OF CDK2/CYCLIN A IN COMPLEX WITH AN OXINDOLE INHIBITOR | | 分子名称: | 4-[(7-OXO-7H-THIAZOLO[5,4-E]INDOL-8-YLMETHYL)-AMINO]-N-PYRIDIN-2-YL-BENZENESULFONAMIDE, CYCLIN A, CYCLIN-DEPENDENT KINASE 2 | | 著者 | Davis, S.T, Benson, B.G, Bramson, H.N, Chapman, D.E, Dickerson, S.H, Dold, K.M, Eberwein, D.J, Edelstein, M, Frye, S.V, Gampe Jr, R.T, Griffin, R.J, Harris, P.A, Hassell, A.M, Holmes, W.D, Hunter, R.N, Knick, V.B, Lackey, K, Lovejoy, B, Luzzio, M.J, Murray, D, Parker, P, Rocque, W.J, Shewchuk, L, Veal, J.M, Walker, D.H, Kuyper, L.K. | | 登録日 | 2000-09-20 | | 公開日 | 2001-01-17 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Prevention of chemotherapy-induced alopecia in rats by CDK inhibitors.
Science, 291, 2001
|
|
1GII
 
 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE CDK4 INHIBITOR | | 分子名称: | 1-(5-OXO-2,3,5,9B-TETRAHYDRO-1H-PYRROLO[2,1-A]ISOINDOL-9-YL)-3-PYRIDIN-2-YL-UREA, CELL DIVISION PROTEIN KINASE 2 | | 著者 | Ikuta, M, Nishimura, S. | | 登録日 | 2001-02-06 | | 公開日 | 2002-02-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystallographic approach to identification of cyclin-dependent kinase 4 (CDK4)-specific inhibitors by using CDK4 mimic CDK2 protein.
J.Biol.Chem., 276, 2001
|
|
1GNF
 
 | | SOLUTION STRUCTURE OF THE N-TERMINAL ZINC FINGER OF MURINE GATA-1, NMR, 25 STRUCTURES | | 分子名称: | TRANSCRIPTION FACTOR GATA-1, ZINC ION | | 著者 | Kowalski, K, Czolij, R, King, G.F, Crossley, M, Mackay, J.P. | | 登録日 | 1998-10-12 | | 公開日 | 1999-06-08 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of the N-terminal zinc finger of GATA-1 reveals a specific binding face for the transcriptional co-factor FOG.
J.Biomol.NMR, 13, 1999
|
|
1GIH
 
 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE CDK4 INHIBITOR | | 分子名称: | 1-(5-OXO-2,3,5,9B-TETRAHYDRO-1H-PYRROLO[2,1-A]ISOINDOL-9-YL)-3-PYRIDIN-2-YL-UREA, CELL DIVISION PROTEIN KINASE 2 | | 著者 | Ikuta, M, Nishimura, S. | | 登録日 | 2001-02-06 | | 公開日 | 2002-02-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystallographic approach to identification of cyclin-dependent kinase 4 (CDK4)-specific inhibitors by using CDK4 mimic CDK2 protein.
J.Biol.Chem., 276, 2001
|
|
1RZY
 
 | | Crystal structure of rabbit Hint complexed with N-ethylsulfamoyladenosine | | 分子名称: | 5'-O-(N-ETHYL-SULFAMOYL)ADENOSINE, Histidine triad nucleotide-binding protein 1 | | 著者 | Krakowiak, A.K, Pace, H.C, Blackburn, G.M, Adams, M, Mekhalfia, A, Kaczmarek, R, Baraniak, J, Stec, W.J, Brenner, C. | | 登録日 | 2003-12-29 | | 公開日 | 2004-03-09 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Biochemical, crystallographic, and mutagenic characterization of hint, the AMP-lysine hydrolase, with novel substrates and inhibitors
J.Biol.Chem., 279, 2004
|
|
2LQN
 
 | |
2LQW
 
 | |
3FVI
 
 | |
1TI7
 
 | | CRYSTAL STRUCTURE OF NMRA, A NEGATIVE TRANSCRIPTIONAL REGULATOR, IN COMPLEX WITH NADP AT 1.7A RESOLUTION | | 分子名称: | CHLORIDE ION, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Lamb, H.K, Leslie, K, Dodds, A.L, Nutley, M, Cooper, A, Johnson, C, Thompson, P, Stammers, D.K, Hawkins, A.R. | | 登録日 | 2004-06-02 | | 公開日 | 2004-06-08 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The negative transcriptional regulator NmrA discriminates between oxidized and reduced dinucleotides.
J.Biol.Chem., 278, 2003
|
|
1QMZ
 
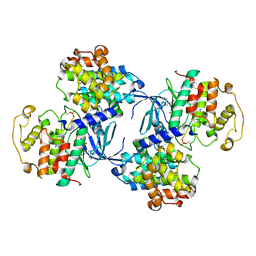 | | PHOSPHORYLATED CDK2-CYCLYIN A-SUBSTRATE PEPTIDE COMPLEX | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CELL DIVISION PROTEIN KINASE 2, G2/MITOTIC-SPECIFIC CYCLIN A, ... | | 著者 | Brown, N.R, Noble, M.E.M, Endicott, J.A, Johnson, L.N. | | 登録日 | 1999-10-11 | | 公開日 | 1999-12-14 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The Structural Basis for Specificity of Substrate and Recruitment Peptides for Cyclin-Dependent Kinases
Nat.Cell Biol., 1, 1999
|
|
2XP4
 
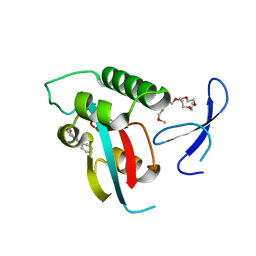 | | DISCOVERY OF CELL-ACTIVE PHENYL-IMIDAZOLE PIN1 INHIBITORS BY STRUCTURE-GUIDED FRAGMENT EVOLUTION | | 分子名称: | 2-phenyl-1H-imidazole-4-carboxylic acid, DODECAETHYLENE GLYCOL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | 著者 | Potter, A, Oldfield, V, Nunns, C, Fromont, C, Ray, S, Northfield, C.J, Bryant, C.J, Scrace, S.F, Robinson, D, Matossova, N, Baker, L, Dokurno, P, Surgenor, A.E, Davis, B.E, Richardson, C.M, Murray, J.B, Moore, J.D. | | 登録日 | 2010-08-25 | | 公開日 | 2011-01-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Discovery of Cell-Active Phenyl-Imidazole Pin1 Inhibitors by Structure-Guided Fragment Evolution.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2X6V
 
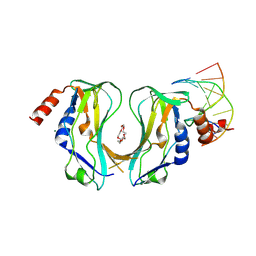 | | Crystal structure of human TBX5 in the DNA-bound and DNA-free form | | 分子名称: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 5'-D(*TP*AP*AP*GP*GP*TP*GP*TP*GP*AP*GP)-3', 5'-D(*TP*CP*TP*CP*AP*CP*AP*CP*CP*TP*TP)-3', ... | | 著者 | Ptchelkine, D, Stirnimann, C.U, Grimm, C, Mueller, C.W. | | 登録日 | 2010-02-22 | | 公開日 | 2010-04-28 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural Basis of Tbx5-DNA Recognition: The T-Box Domain in its DNA-Bound and -Unbound Form.
J.Mol.Biol., 400, 2010
|
|
2XP6
 
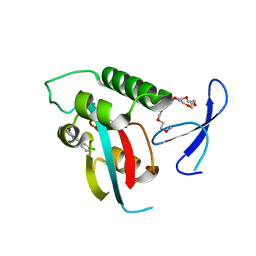 | | DISCOVERY OF CELL-ACTIVE PHENYL-IMIDAZOLE PIN1 INHIBITORS BY STRUCTURE-GUIDED FRAGMENT EVOLUTION | | 分子名称: | 2-(3-CHLORO-PHENYL)-5-METHYL-1H-IMIDAZOLE-4-CARBOXYLIC ACID, DODECAETHYLENE GLYCOL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | 著者 | Potter, A, Oldfield, V, Nunns, C, Fromont, C, Ray, S, Northfield, C.J, Bryant, C.J, Scrace, S.F, Robinson, D, Matossova, N, Baker, L, Dokurno, P, Surgenor, A.E, Davis, B.E, Richardson, C.M, Murray, J.B, Moore, J.D. | | 登録日 | 2010-08-25 | | 公開日 | 2011-01-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Discovery of Cell-Active Phenyl-Imidazole Pin1 Inhibitors by Structure-Guided Fragment Evolution.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
1R36
 
 | | NMR-based structure of autoinhibited murine Ets-1 deltaN301 | | 分子名称: | C-ets-1 protein | | 著者 | Lee, G.M, Donaldson, L.W, Pufall, M.A, Kang, H.-S, Pot, I, Graves, B.J, McIntosh, L.P. | | 登録日 | 2003-09-30 | | 公開日 | 2004-11-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Structural and Dynamic Basis of Ets-1 DNA Binding Autoinhibition
J.Biol.Chem., 280, 2005
|
|
2XPB
 
 | | DISCOVERY OF CELL-ACTIVE PHENYL-IMIDAZOLE PIN1 INHIBITORS BY STRUCTURE-GUIDED FRAGMENT EVOLUTION | | 分子名称: | 5-[BENZYL(METHYL)CARBAMOYL]-2-(3-CHLOROPHENYL)-1H-IMIDAZOLE-4-CARBOXYLIC ACID, DODECAETHYLENE GLYCOL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | 著者 | Potter, A, Oldfield, V, Nunns, C, Fromont, C, Ray, S, Northfield, C.J, Bryant, C.J, Scrace, S.F, Robinson, D, Matossova, N, Baker, L, Dokurno, P, Surgenor, A.E, Davis, B.E, Richardson, C.M, Murray, J.B, Moore, J.D. | | 登録日 | 2010-08-25 | | 公開日 | 2011-01-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Discovery of Cell-Active Phenyl-Imidazole Pin1 Inhibitors by Structure-Guided Fragment Evolution.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
1RG8
 
 | |
2XYI
 
 | | Crystal Structure of Nurf55 in complex with a H4 peptide | | 分子名称: | DI(HYDROXYETHYL)ETHER, HISTONE H4, PROBABLE HISTONE-BINDING PROTEIN CAF1, ... | | 著者 | Stirnimann, C.U, Nowak, A.J, Mueller, C.W. | | 登録日 | 2010-11-17 | | 公開日 | 2011-05-04 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Chromatin-Modifying Complex Component Nurf55/P55 Associates with Histones H3, H4 and Polycomb Repressive Complex 2 Subunit Su(Z)12 Through Partially Overlapping Binding Sites.
J.Biol.Chem., 286, 2011
|
|
3FVJ
 
 | |
2XP7
 
 | | DISCOVERY OF CELL-ACTIVE PHENYL-IMIDAZOLE PIN1 INHIBITORS BY STRUCTURE-GUIDED FRAGMENT EVOLUTION | | 分子名称: | 2-PHENYL-1H-IMIDAZOLE-4,5-DICARBOXYLIC ACID, DODECAETHYLENE GLYCOL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | 著者 | Potter, A, Oldfield, V, Nunns, C, Fromont, C, Ray, S, Northfield, C.J, Bryant, C.J, Scrace, S.F, Robinson, D, Matossova, N, Baker, L, Dokurno, P, Surgenor, A.E, Davis, B.E, Richardson, C.M, Murray, J.B, Moore, J.D. | | 登録日 | 2010-08-25 | | 公開日 | 2011-01-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Discovery of Cell-Active Phenyl-Imidazole Pin1 Inhibitors by Structure-Guided Fragment Evolution.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
1SAN
 
 | |
2XP8
 
 | | DISCOVERY OF CELL-ACTIVE PHENYL-IMIDAZOLE PIN1 INHIBITORS BY STRUCTURE-GUIDED FRAGMENT EVOLUTION | | 分子名称: | 4-(MORPHOLIN-4-YLCARBONYL)-2-PHENYL-1H-IMIDAZOLE-5-CARBOXYLIC ACID, DODECAETHYLENE GLYCOL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | 著者 | Potter, A, Oldfield, V, Nunns, C, Fromont, C, Ray, S, Northfield, C.J, Bryant, C.J, Scrace, S.F, Robinson, D, Matossova, N, Baker, L, Dokurno, P, Surgenor, A.E, Davis, B.E, Richardson, C.M, Murray, J.B, Moore, J.D. | | 登録日 | 2010-08-25 | | 公開日 | 2011-01-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Discovery of Cell-Active Phenyl-Imidazole Pin1 Inhibitors by Structure-Guided Fragment Evolution.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
