5YTS
 
 | |
5YTV
 
 | |
5XV9
 
 | |
1H95
 
 | | Solution structure of the single-stranded DNA-binding Cold Shock Domain (CSD) of human Y-box protein 1 (YB1) determined by NMR (10 lowest energy structures) | | 分子名称: | Y-BOX BINDING PROTEIN | | 著者 | Kloks, C.P.A.M, Spronk, C.A.E.M, Hoffmann, A, Vuister, G.W, Grzesiek, S, Hilbers, C.W. | | 登録日 | 2001-02-23 | | 公開日 | 2002-02-21 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Solution Structure and DNA-Binding Properties of the Cold-Shock Domain of the Human Y-Box Protein Yb-1.
J.Mol.Biol., 316, 2002
|
|
1I5F
 
 | | BACILLUS CALDOLYTICUS COLD-SHOCK PROTEIN MUTANTS TO STUDY DETERMINANTS OF PROTEIN STABILITY | | 分子名称: | COLD-SHOCK PROTEIN CSPB, SODIUM ION | | 著者 | Delbrueck, H, Mueller, U, Perl, D, Schmid, F.X, Heinemann, U. | | 登録日 | 2001-02-27 | | 公開日 | 2001-11-07 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal structures of mutant forms of the Bacillus caldolyticus cold shock protein differing in thermal stability.
J.Mol.Biol., 313, 2001
|
|
1G6P
 
 | | SOLUTION NMR STRUCTURE OF THE COLD SHOCK PROTEIN FROM THE HYPERTHERMOPHILIC BACTERIUM THERMOTOGA MARITIMA | | 分子名称: | COLD SHOCK PROTEIN TMCSP | | 著者 | Kremer, W, Schuler, B, Harrieder, S, Geyer, M, Gronwald, W, Welker, C, Jaenicke, R, Kalbitzer, H.R. | | 登録日 | 2000-11-07 | | 公開日 | 2001-11-07 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR structure of the cold-shock protein from the hyperthermophilic bacterium Thermotoga maritima.
Eur.J.Biochem., 268, 2001
|
|
1HZ9
 
 | | BACILLUS CALDOLYTICUS COLD-SHOCK PROTEIN MUTANTS TO STUDY DETERMINANTS OF PROTEIN STABILITY | | 分子名称: | COLD SHOCK PROTEIN CSPB | | 著者 | Delbrueck, H, Mueller, U, Perl, D, Schmid, F.X, Heinemann, U. | | 登録日 | 2001-01-24 | | 公開日 | 2001-11-07 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structures of mutant forms of the Bacillus caldolyticus cold shock protein differing in thermal stability.
J.Mol.Biol., 313, 2001
|
|
1HZA
 
 | | BACILLUS CALDOLYTICUS COLD-SHOCK PROTEIN MUTANTS TO STUDY DETERMINANTS OF PROTEIN STABILITY | | 分子名称: | COLD SHOCK PROTEIN CSPB | | 著者 | Delbrueck, H, Mueller, U, Perl, D, Schmid, F.X, Heinemann, U. | | 登録日 | 2001-01-24 | | 公開日 | 2001-11-07 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structures of mutant forms of the Bacillus caldolyticus cold shock protein differing in thermal stability.
J.Mol.Biol., 313, 2001
|
|
1HZB
 
 | | BACILLUS CALDOLYTICUS COLD-SHOCK PROTEIN MUTANTS TO STUDY DETERMINANTS OF PROTEIN STABILITY | | 分子名称: | COLD SHOCK PROTEIN CSPB, SODIUM ION | | 著者 | Delbrueck, H, Mueller, U, Perl, D, Schmid, F.X, Heinemann, U. | | 登録日 | 2001-01-24 | | 公開日 | 2001-11-07 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.28 Å) | | 主引用文献 | Crystal structures of mutant forms of the Bacillus caldolyticus cold shock protein differing in thermal stability.
J.Mol.Biol., 313, 2001
|
|
1HZC
 
 | | BACILLUS CALDOLYTICUS COLD-SHOCK PROTEIN MUTANTS TO STUDY DETERMINANTS OF PROTEIN STABILITY | | 分子名称: | COLD SHOCK PROTEIN CSPB, SODIUM ION | | 著者 | Delbrueck, H, Mueller, U, Perl, D, Schmid, F.X, Heinemann, U. | | 登録日 | 2001-01-24 | | 公開日 | 2001-11-07 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | Crystal structures of mutant forms of the Bacillus caldolyticus cold shock protein differing in thermal stability.
J.Mol.Biol., 313, 2001
|
|
2I5L
 
 | |
2I5M
 
 | |
2L15
 
 | | Solution Structure of Cold Shock Protein CspA Using Combined NMR and CS-Rosetta method | | 分子名称: | Cold shock protein CspA | | 著者 | Tang, Y, Schneider, W.M, Shen, Y, Raman, S, Inouye, M, Baker, D, Roth, M.J, Montelione, G.T. | | 登録日 | 2010-07-22 | | 公開日 | 2010-09-15 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Fully automated high-quality NMR structure determination of small (2)H-enriched proteins.
J Struct Funct Genomics, 11, 2010
|
|
2LSS
 
 | | Solution structure of the R. rickettsii cold shock-like protein | | 分子名称: | Cold shock-like protein | | 著者 | Veldkamp, C.T, Peterson, F.C, Gerarden, K.P, Fuchs, A.M, Koch, J.M, Mueller, M.M. | | 登録日 | 2012-05-04 | | 公開日 | 2012-05-16 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the cold-shock-like protein from Rickettsia rickettsii.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
2LXK
 
 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for cold shock protein, LmCsp | | 分子名称: | Cold shock-like protein CspLA | | 著者 | Lee, J, Jeong, K, Kim, Y. | | 登録日 | 2012-08-27 | | 公開日 | 2013-08-07 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural and Dynamic Features of Cold-Shock Proteins of Listeriamonocytogenes, a Psychrophilic Bacterium
Biochemistry, 52, 2013
|
|
2LXJ
 
 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for cold shock protein, LmCsp with dT7 | | 分子名称: | Cold shock-like protein CspLA | | 著者 | Lee, J, Jeong, K, Kim, Y. | | 登録日 | 2012-08-27 | | 公開日 | 2013-08-07 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural and Dynamic Features of Cold-Shock Proteins of Listeriamonocytogenes, a Psychrophilic Bacterium
Biochemistry, 52, 2013
|
|
2K5N
 
 | | Solution NMR Structure of the N-Terminal Domain of Protein ECA1580 from Erwinia carotovora, Northeast Structural Genomics Consortium Target EwR156A | | 分子名称: | Putative cold-shock protein | | 著者 | Mills, J.L, Eletsky, A, Zhang, Q, Lee, D, Jiang, M, Ciccosanti, C, Xiao, R, Lui, J, Everett, J.K, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2008-06-30 | | 公開日 | 2008-08-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Structure of the Putative Cold Shock Protein from Erwinia carotovora: Northeast Structural Genomics Consortium Target EwR156a
To be Published
|
|
2KCM
 
 | | Solution NMR structure of the N-terminal OB-domain of SO_1732 from Shewanella oneidensis. Northeast Structural Genomics Consortium Target SoR210A. | | 分子名称: | Cold shock domain family protein | | 著者 | Ramelot, T.A, Maglaqui, M, Jiang, M, Ciccosanti, C, Xiao, R, Lui, J, Everett, J.K, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2008-12-23 | | 公開日 | 2009-02-10 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR structure of the N-terminal OB-domain of SO_1732 from Shewanella oneidensis. Northeast Structural Genomics Consortium Target SoR210A.
To be Published
|
|
8H1I
 
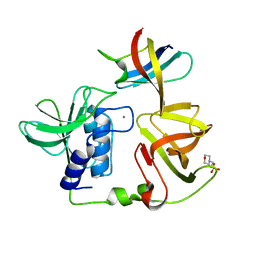 | | Crystal structure of PlyGRCS, a bacteriophage Endolysin in complex with Cold shock protein C | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Cold shock-like protein CspC, ... | | 著者 | Padmanabhan, B, Gopinatha, K, Mandal, M, Saranya, G, Sudhagar, B. | | 登録日 | 2022-10-03 | | 公開日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of PlyGRCS, a bacteriophage Endolysin in complex with Cold shock protein C
To Be Published
|
|
7OII
 
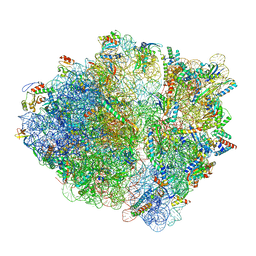 | | CspA-70 cotranslational folding intermediate 2 | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Agirrezabala, X, Samatova, E, Macher, M, Liutkute, M, Gil-Carton, D, Novacek, J, Valle, M, Rodnina, M.V. | | 登録日 | 2021-05-11 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | A switch from alpha-helical to beta-strand conformation during co-translational protein folding.
Embo J., 41, 2022
|
|
4QQB
 
 | | Structural basis for the assembly of the SXL-UNR translation regulatory complex | | 分子名称: | Protein sex-lethal, Upstream of N-ras, isoform A, ... | | 著者 | Hennig, J, Popowicz, G.M, Sattler, M. | | 登録日 | 2014-06-27 | | 公開日 | 2014-09-03 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis for the assembly of the Sxl-Unr translation regulatory complex.
Nature, 515, 2014
|
|
5UDZ
 
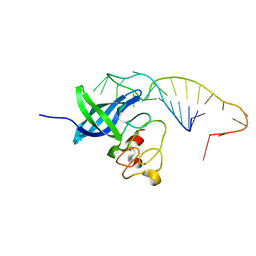 | |
7ZHR
 
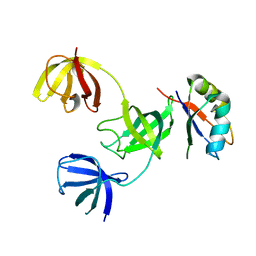 | |
8OST
 
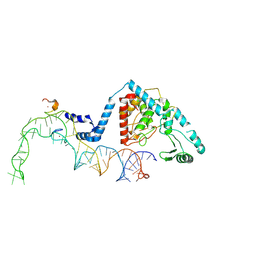 | | Structure of human terminal uridylyltransferase 4 (TUT4, ZCCHC11) in complex with pre-let7g miRNA and Lin28A | | 分子名称: | Protein lin-28 homolog A, Terminal uridylyltransferase 4, ZINC ION, ... | | 著者 | Gilbert, R.J, Yi, G, Ye, M. | | 登録日 | 2023-04-20 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-09-25 | | 実験手法 | ELECTRON MICROSCOPY (3.69 Å) | | 主引用文献 | Structural basis for activity switching in polymerases determining the fate of let-7 pre-miRNAs.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8OPS
 
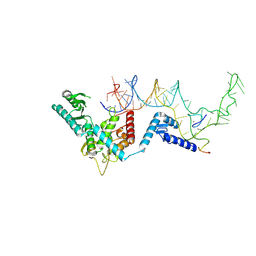 | | Human terminal uridylyltransferase 7 (TUT7/ZCCHC6) bound with pre-let7g miRNA and Lin28A - complex 1 | | 分子名称: | Protein lin-28 homolog A, RNA (71-MER) Let7g, Terminal uridylyltransferase 7, ... | | 著者 | Yi, G, Ye, M, Gilbert, R.J. | | 登録日 | 2023-04-08 | | 公開日 | 2024-07-24 | | 最終更新日 | 2024-09-25 | | 実験手法 | ELECTRON MICROSCOPY (3.82 Å) | | 主引用文献 | Structural basis for activity switching in polymerases determining the fate of let-7 pre-miRNAs.
Nat.Struct.Mol.Biol., 31, 2024
|
|
