3A4P
 
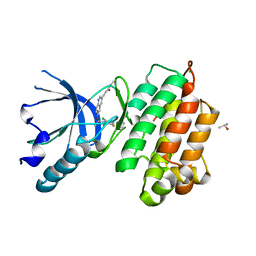 | | human c-MET kinase domain complexed with 6-benzyloxyquinoline inhibitor | | 分子名称: | (2E)-3-{6-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]quinolin-3-yl}-N-methylprop-2-enamide, CHLORIDE ION, Hepatocyte growth factor receptor, ... | | 著者 | Fukami, T.A, Kadono, S, Yamamuro, M, Matsuura, T. | | 登録日 | 2009-07-13 | | 公開日 | 2010-02-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | Discovery of 6-benzyloxyquinolines as c-Met selective kinase inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
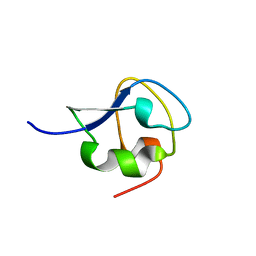
7Q3V
 
 | |
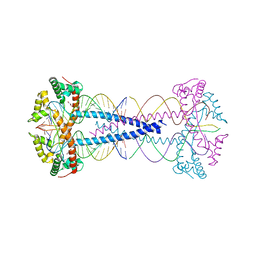
7ZIE
 
 | |
8E0G
 
 | |
9IYK
 
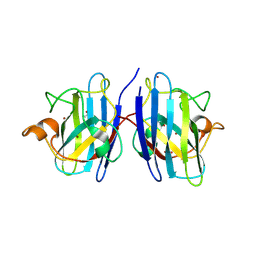 | |
5DOW
 
 | |
9F3Y
 
 | |
9F3X
 
 | |
8A8L
 
 | | Crystal structure of a staphylococcal orthologue of CYP134A1 (CYPX) in complex with a heme-coordinated fragment | | 分子名称: | 6-methoxy-2,3,4,9-tetrahydro-1H-pyrido[3,4-b]indole, Cytochrome P450 protein, GLYCEROL, ... | | 著者 | Snee, M, Katariya, M, Levy, C. | | 登録日 | 2022-06-23 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Crystal structure of a staphylococcal orthologue of CYP134A1 (CYPX) in complex with a heme-coordinated fragment
To Be Published
|
|
8A6W
 
 | |
8A9P
 
 | | Crystal structure of CYP142 from Mycobacterium tuberculosis in complex with a fragment | | 分子名称: | (3-phenyl-1,2,4-oxadiazol-5-yl)methanamine, BROMIDE ION, CHLORIDE ION, ... | | 著者 | Snee, M, Katariya, M, Levy, C, Leys, D. | | 登録日 | 2022-06-29 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Crystal structure of CYP142 from Mycobacterium tuberculosis in complex with a fragment
To Be Published
|
|
6ZFW
 
 | | X-ray structure of the soluble N-terminal domain of T. cruzi PEX-14 | | 分子名称: | ACETATE ION, BETA-MERCAPTOETHANOL, GLYCEROL, ... | | 著者 | Softley, C.A, Ostertag, M.O, Sattler, M, Popowicz, G.P. | | 登録日 | 2020-06-18 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Deep learning model predicts water interaction sites on the surface of proteins using limited-resolution data.
Chem.Commun.(Camb.), 56, 2020
|
|
6ZK0
 
 | | 1.47A human IMPase with ebselen | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Inositol monophosphatase 1, ... | | 著者 | Bax, B.D, Fenn, G.D. | | 登録日 | 2020-06-29 | | 公開日 | 2020-09-23 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Crystallization and structure of ebselen bound to Cys141 of human inositol monophosphatase.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
7XQ3
 
 | |
7XPW
 
 | | Structure of OhTRP14 | | 分子名称: | Thioredoxin domain-containing protein 17 | | 著者 | Wang, S.Q, Huang, S.Q. | | 登録日 | 2022-05-05 | | 公開日 | 2022-10-12 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Structural insights into the redox regulation of Oncomelania hupensis TRP14 and its potential role in the snail host response to parasite invasion.
Fish Shellfish Immunol., 128, 2022
|
|
8ASO
 
 | |
8ASM
 
 | |
8F8O
 
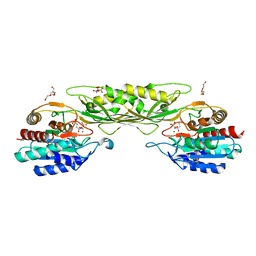 | | Crystal Structure of the Succinyl-diaminopimelate Desuccinylase (DapE) from Acinetobacter baumannii in complex with Succinic and L-Lactic Acids | | 分子名称: | (2S)-2-HYDROXYPROPANOIC ACID, CITRIC ACID, SUCCINIC ACID, ... | | 著者 | Minasov, G, Shuvalova, L, Brunzelle, J.S, Dubrovska, I, Pshenychnyi, S, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-11-22 | | 公開日 | 2022-11-30 | | 最終更新日 | 2025-09-24 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Biochemical and Structural Analysis of the Bacterial Enzyme Succinyl-Diaminopimelate Desuccinylase (DapE) from Acinetobacter baumannii.
Acs Omega, 9, 2024
|
|
9JUR
 
 | |
9JS5
 
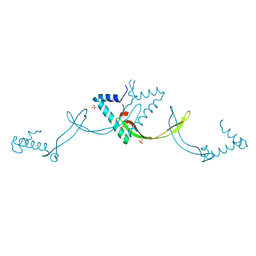 | | Crystal structure of the ASFV-derived histone-like protein pA104R | | 分子名称: | 1,2-ETHANEDIOL, SULFATE ION, Viral histone-like protein | | 著者 | Li, Q, Shao, H, Yi, D, Cen, S. | | 登録日 | 2024-09-30 | | 公開日 | 2025-04-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Thonningianin A disrupts pA104R-DNA binding and inhibits African swine fever virus replication.
Emerg Microbes Infect, 14, 2025
|
|
8UEL
 
 | |
6MV1
 
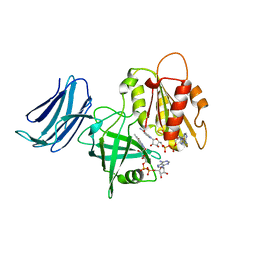 | | 2.15A resolution structure of the CS-b5R domains of human Ncb5or (NAD+ form) | | 分子名称: | Cytochrome b5 reductase 4, FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Lovell, S, Mehzabeen, N, Battaile, K.P, Benson, D.R, Cooper, A, Gao, P, Zhu, H. | | 登録日 | 2018-10-24 | | 公開日 | 2019-07-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystal structures of the naturally fused CS and cytochrome b5reductase (b5R) domains of Ncb5or reveal an expanded CS fold, extensive CS-b5R interactions and productive binding of the NAD(P)+nicotinamide ring.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6MV2
 
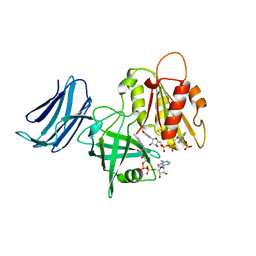 | | 2.05A resolution structure of the CS-b5R domains of human Ncb5or (NADP+ form) | | 分子名称: | Cytochrome b5 reductase 4, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Lovell, S, Mehzabeen, N, Battaile, K.P, Benson, D.R, Cooper, A, Gao, P, Zhu, H. | | 登録日 | 2018-10-24 | | 公開日 | 2019-07-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Crystal structures of the naturally fused CS and cytochrome b5reductase (b5R) domains of Ncb5or reveal an expanded CS fold, extensive CS-b5R interactions and productive binding of the NAD(P)+nicotinamide ring.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
7VUM
 
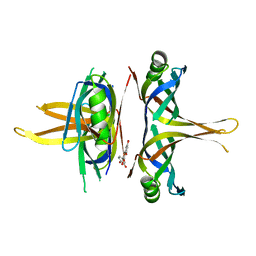 | | Crystal structure of SSB complexed with que | | 分子名称: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, Single-stranded DNA-binding protein | | 著者 | Lin, E.S, Huang, Y.H, Huang, C.Y. | | 登録日 | 2021-11-03 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.319 Å) | | 主引用文献 | A Complexed Crystal Structure of a Single-Stranded DNA-Binding Protein with Quercetin and the Structural Basis of Flavonol Inhibition Specificity.
Int J Mol Sci, 23, 2022
|
|
3F5E
 
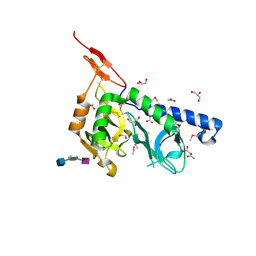 | | Crystal structure of Toxoplasma gondii micronemal protein 1 bound to 2'F-3'SiaLacNAc1-3 | | 分子名称: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Garnett, J.A, Liu, Y, Feizi, T, Matthews, S.J. | | 登録日 | 2008-11-03 | | 公開日 | 2009-07-28 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Detailed insights from microarray and crystallographic studies into carbohydrate recognition by microneme protein 1 (MIC1) of Toxoplasma gondii.
Protein Sci., 18, 2009
|
|
