3LJ9
 
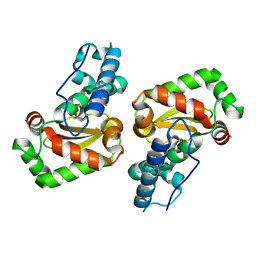 | | X-ray structure of the iron superoxide dismutase from pseudoalteromonas haloplanktis in complex with sodium azide | | 分子名称: | AZIDE ION, FE (III) ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, ... | | 著者 | Merlino, A, Russo Krauss, I, Rossi, B, Conte, M, Vergara, A, Sica, F. | | 登録日 | 2010-01-26 | | 公開日 | 2010-09-08 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure and flexibility in cold-adapted iron superoxide dismutases: the case of the enzyme isolated from Pseudoalteromonas haloplanktis.
J.Struct.Biol., 172, 2010
|
|
2H6A
 
 | | Crystal structure of the zinc-beta-lactamase L1 from Stenotrophomonas maltophilia (mono zinc form) | | 分子名称: | Metallo-beta-lactamase L1, SULFATE ION, ZINC ION | | 著者 | Nauton, L, Garau, G, Kahn, R, Dideberg, O. | | 登録日 | 2006-05-31 | | 公開日 | 2007-04-17 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural insights into the design of inhibitors for the L1 metallo-beta-lactamase from Stenotrophomonas maltophilia.
J.Mol.Biol., 375, 2008
|
|
7X40
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 8A10 (classified from CVB1 mature virion in complex with 8A10 and 2E6) | | 分子名称: | 8A10 heavy chain, 8A10 light chain, Capsid protein VP4, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-03-01 | | 公開日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3.02 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X46
 
 | | Cryo-EM structure of Coxsackievirus B1 A-particle in complex with nAb 2E6 (classified from CVB1 mature virion in complex with 8A10 and 2E6) | | 分子名称: | 2E6 heavy chain, 2E6 light chain, VP2, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-03-02 | | 公開日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3.85 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X47
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb 2E6 (classified from CVB1 mature virion in complex with 8A10 and 2E6) | | 分子名称: | 2E6 heavy chain, 2E6 light chain, Genome polyprotein, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-03-02 | | 公開日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3.66 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
1LGR
 
 | |
7X3C
 
 | | Cryo-EM structure of Coxsackievirus B1 muture virion in complex with nAbs 8A10 and 5F5 (CVB1-M:8A10:5F5) | | 分子名称: | 5F5 heavy chain, 5F5 light chain, 8A10 heavy chain, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-02-28 | | 公開日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3.03 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X4K
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb 9A3 (classified from CVB1 mature virion in complex with 8A10 and 9A3) | | 分子名称: | 9A3 heavy chain, 9A3 light chain, Genome polyprotein, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-03-02 | | 公開日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3.82 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X4M
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 8A10 (classified from CVB1 mature virion in complex with 8A10, 2E6 and 9A3) | | 分子名称: | 8A10 heavy chain, 8A10 light chain, Capsid protein VP4, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-03-02 | | 公開日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3.34 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
1PN6
 
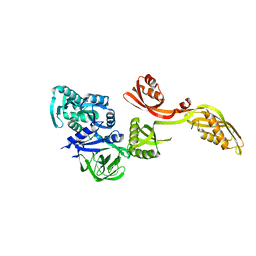 | | Domain-wise fitting of the crystal structure of T.thermophilus EF-G into the low resolution map of the release complex.Puromycin.EFG.GDPNP of E.coli 70S ribosome. | | 分子名称: | Elongation factor G | | 著者 | Valle, M, Zavialov, A, Sengupta, J, Rawat, U, Ehrenberg, M, Frank, J. | | 登録日 | 2003-06-12 | | 公開日 | 2003-07-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (10.8 Å) | | 主引用文献 | Locking and Unlocking of Ribosomal Motions
Cell(Cambridge,Mass.), 114, 2003
|
|
1PS5
 
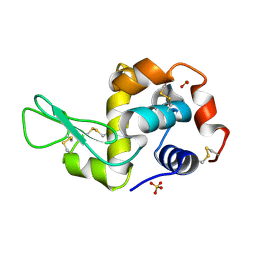 | | STRUCTURE OF THE MONOCLINIC C2 FORM OF HEN EGG-WHITE LYSOZYME AT 2.0 ANGSTROMS RESOLUTION | | 分子名称: | Lysozyme C, SULFATE ION | | 著者 | Majeed, S, Ofek, G, Belachew, A, Huang, C, Zhou, T, Kwong, P.D. | | 登録日 | 2003-06-20 | | 公開日 | 2003-09-09 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Enhancing Protein Crystallization through Precipitant Synergy
Structure, 11, 2003
|
|
2XUA
 
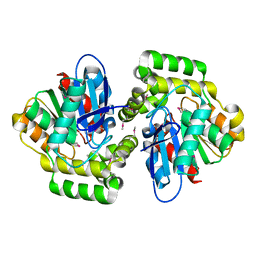 | | Crystal structure of the enol-lactonase from Burkholderia xenovorans LB400 | | 分子名称: | 3-OXOADIPATE ENOL-LACTONASE, LAEVULINIC ACID | | 著者 | Bains, J, Kaufman, L, Farnell, B, Boulanger, M.J. | | 登録日 | 2010-10-17 | | 公開日 | 2011-01-26 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A Product Analog Bound Form of 3-Oxoadipate-Enol- Lactonase (Pcad) Reveals a Multifunctional Role for the Divergent CAP Domain.
J.Mol.Biol., 406, 2011
|
|
1ZN1
 
 | | Coordinates of RRF fitted into Cryo-EM map of the 70S post-termination complex | | 分子名称: | 30S ribosomal protein S12, Ribosome recycling factor, ribosomal 16S RNA, ... | | 著者 | Gao, N, Zavialov, A.V, Li, W, Sengupta, J, Valle, M, Gursky, R.P, Ehrenberg, M, Frank, J. | | 登録日 | 2005-05-11 | | 公開日 | 2005-06-14 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (14.1 Å) | | 主引用文献 | Mechanism for the disassembly of the posttermination complex inferred from cryo-EM studies.
Mol.Cell, 18, 2005
|
|
1NNP
 
 | | X-ray structure of the GluR2 ligand-binding core (S1S2J) in complex with (S)-ATPA at 1.9 A resolution. Crystallization without zinc ions. | | 分子名称: | 3-(5-TERT-BUTYL-3-OXIDOISOXAZOL-4-YL)-L-ALANINATE, Glutamate receptor 2, SULFATE ION | | 著者 | Lunn, M.L, Hogner, A, Stensbol, T.B, Gouaux, E, Egebjerg, J, Kastrup, J.S. | | 登録日 | 2003-01-14 | | 公開日 | 2003-03-11 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Three-Dimensional Structure of the Ligand-Binding
Core of GluR2 in Complex with the Agonist (S)-ATPA:
Implications for Receptor Subunit Selectivity.
J.Med.Chem., 46, 2003
|
|
2XPZ
 
 | | Structure of native yeast LTA4 hydrolase | | 分子名称: | (R,R)-2,3-BUTANEDIOL, LEUKOTRIENE A-4 HYDROLASE, POLYETHYLENE GLYCOL (N=34), ... | | 著者 | Helgstrand, C, Hasan, M, Usyal, H, Haeggstrom, J.Z, Thunnissen, M.M.G.M. | | 登録日 | 2010-08-31 | | 公開日 | 2010-12-29 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | A Leukotriene A(4) Hydrolase-Related Aminopeptidase from Yeast Undergoes Induced Fit Upon Inhibitor Binding.
J.Mol.Biol., 406, 2011
|
|
7YWF
 
 | | Monocot chimeric jacalin JAC1 from Oryza sativa: dirigent domain with bound galactobiose | | 分子名称: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Dirigent protein, ... | | 著者 | Huwa, N, Classen, T, Weiergraeber, O.H. | | 登録日 | 2022-02-13 | | 公開日 | 2022-05-04 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The Crystal Structure of the Defense Conferring Rice Protein Os JAC1 Reveals a Carbohydrate Binding Site on the Dirigent-like Domain.
Biomolecules, 12, 2022
|
|
7YWE
 
 | | Monocot chimeric jacalin JAC1 from Oryza sativa: dirigent domain (crystal form 2) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Dirigent protein, PHOSPHATE ION | | 著者 | Huwa, N, Classen, T, Weiergraeber, O.H. | | 登録日 | 2022-02-13 | | 公開日 | 2022-05-04 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | The Crystal Structure of the Defense Conferring Rice Protein Os JAC1 Reveals a Carbohydrate Binding Site on the Dirigent-like Domain.
Biomolecules, 12, 2022
|
|
1LPB
 
 | | THE 2.46 ANGSTROMS RESOLUTION STRUCTURE OF THE PANCREATIC LIPASE COLIPASE COMPLEX INHIBITED BY A C11 ALKYL PHOSPHONATE | | 分子名称: | CALCIUM ION, COLIPASE, LIPASE, ... | | 著者 | Egloff, M.-P, Van Tilbeurgh, H, Cambillau, C. | | 登録日 | 1994-08-19 | | 公開日 | 1994-11-01 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | The 2.46 A resolution structure of the pancreatic lipase-colipase complex inhibited by a C11 alkyl phosphonate.
Biochemistry, 34, 1995
|
|
7YWW
 
 | | Monocot chimeric jacalin JAC1 from Oryza sativa: lectin domain (crystal form 2) | | 分子名称: | Dirigent protein, GLYCEROL, IODIDE ION, ... | | 著者 | Huwa, N, Classen, T, Weiergraeber, O.H. | | 登録日 | 2022-02-14 | | 公開日 | 2022-05-04 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | The Crystal Structure of the Defense Conferring Rice Protein Os JAC1 Reveals a Carbohydrate Binding Site on the Dirigent-like Domain.
Biomolecules, 12, 2022
|
|
7YWG
 
 | |
3ZUW
 
 | | Photosynthetic Reaction Centre Mutant with TYR L128 replaced with HIS | | 分子名称: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | 著者 | Gibasiewicz, K, Pajzderska, M, Potter, J.A, Fyfe, P.K, Dobek, A, Brettel, K, Jones, M.R. | | 登録日 | 2011-07-20 | | 公開日 | 2011-11-16 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Mechanism of Recombination of the P(+)H(A)(-) Radical Pair in Mutant Rhodobacter Sphaeroides Reaction Centers with Modified Free Energy Gaps between P(+)B(A)(-) and P(+)H(A)(-).
J Phys Chem B, 115, 2011
|
|
1DJB
 
 | |
7ZAL
 
 | |
3LIO
 
 | | X-ray structure of the iron superoxide dismutase from pseudoalteromonas haloplanktis (crystal form I) | | 分子名称: | FE (III) ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, iron superoxide dismutase | | 著者 | Merlino, A, Russo Krauss, I, Rossi, B, Conte, M, Vergara, A, Sica, F. | | 登録日 | 2010-01-25 | | 公開日 | 2010-09-08 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure and flexibility in cold-adapted iron superoxide dismutases: the case of the enzyme isolated from Pseudoalteromonas haloplanktis.
J.Struct.Biol., 172, 2010
|
|
1DXH
 
 | | Catabolic ornithine carbamoyltransferase from Pseudomonas aeruginosa | | 分子名称: | ORNITHINE CARBAMOYLTRANSFERASE, SULFATE ION | | 著者 | Sainz, G, Vicat, J, Kahn, R, Duee, E, Tricot, C, Stalon, V, Dideberg, O. | | 登録日 | 2000-01-05 | | 公開日 | 2001-01-12 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of the Allosteric Active Form of Catabolic Ornithine Carbamoyltransferase from Pseudomonas Aeruginosa
To be Published
|
|
