5QKK
 
 | |
5QKZ
 
 | |
5QLD
 
 | |
5QLT
 
 | |
5QM8
 
 | |
5QMN
 
 | |
5QN2
 
 | |
5QNH
 
 | |
5QNZ
 
 | |
5QOE
 
 | |
5Q8F
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 224) | | 分子名称: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | 著者 | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | 登録日 | 2017-05-25 | | 公開日 | 2018-08-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | PanDDA analysis group deposition
To Be Published
|
|
5Q8S
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 238) | | 分子名称: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | 著者 | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | 登録日 | 2017-05-25 | | 公開日 | 2018-08-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | PanDDA analysis group deposition
To Be Published
|
|
8DKZ
 
 | |
5Q9A
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 256) | | 分子名称: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | 著者 | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | 登録日 | 2017-05-25 | | 公開日 | 2018-08-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | PanDDA analysis group deposition
To Be Published
|
|
5Q9P
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 271) | | 分子名称: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | 著者 | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | 登録日 | 2017-05-25 | | 公開日 | 2018-08-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | PanDDA analysis group deposition
To Be Published
|
|
8DJJ
 
 | |
5QA8
 
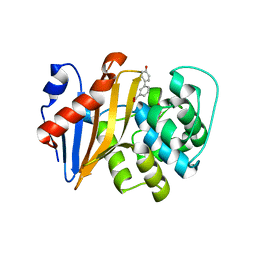 | | OXA-48 IN COMPLEX WITH COMPOUND 4c | | 分子名称: | 3-(4-hydroxyphenyl)benzoic acid, Beta-lactamase, CHLORIDE ION | | 著者 | Lund, B.A, Leiros, H.K.S. | | 登録日 | 2017-07-11 | | 公開日 | 2018-01-10 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A focused fragment library targeting the antibiotic resistance enzyme - Oxacillinase-48: Synthesis, structural evaluation and inhibitor design.
Eur J Med Chem, 145, 2018
|
|
5QAK
 
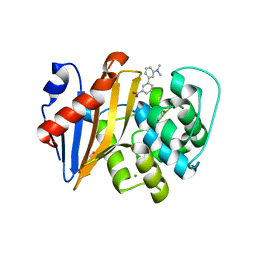 | | OXA-48 IN COMPLEX WITH COMPOUND 14 | | 分子名称: | 1,2-ETHANEDIOL, 3-[3-(dimethylamino)phenyl]benzoic acid, Beta-lactamase, ... | | 著者 | Lund, B.A, Leiros, H.K.S. | | 登録日 | 2017-07-11 | | 公開日 | 2018-01-10 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A focused fragment library targeting the antibiotic resistance enzyme - Oxacillinase-48: Synthesis, structural evaluation and inhibitor design.
Eur J Med Chem, 145, 2018
|
|
8DI3
 
 | |
5QB1
 
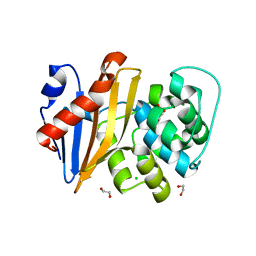 | | OXA-48 IN COMPLEX WITH COMPOUND 36 | | 分子名称: | 1,2-ETHANEDIOL, 3,5-bis(3-acetamidophenyl)benzoic acid, Beta-lactamase, ... | | 著者 | Lund, B.A, Leiros, H.K.S. | | 登録日 | 2017-07-11 | | 公開日 | 2018-01-10 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A focused fragment library targeting the antibiotic resistance enzyme - Oxacillinase-48: Synthesis, structural evaluation and inhibitor design.
Eur J Med Chem, 145, 2018
|
|
8DKK
 
 | |
5QHI
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human FAM83B in complex with FMOPL000271a | | 分子名称: | 1,2-ETHANEDIOL, IODIDE ION, Protein FAM83B, ... | | 著者 | Pinkas, D.M, Bufton, J.C, Fox, A.E, Talon, R, Krojer, T, Douangamath, A, Collins, P, Zhang, R, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Bullock, A.N. | | 登録日 | 2018-05-18 | | 公開日 | 2018-12-19 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
8DK8
 
 | |
5QI0
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000352a | | 分子名称: | CHLORIDE ION, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 14, ... | | 著者 | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | 登録日 | 2018-05-21 | | 公開日 | 2019-04-10 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QIW
 
 | | Covalent fragment group deposition -- Crystal Structure of OUTB2 in complex with PCM-0102660 | | 分子名称: | N-[(E)-(4-methylphenyl)methylidene]acetamide, UNKNOWN LIGAND, Ubiquitin thioesterase OTUB2 | | 著者 | Sethi, R, Douangamath, A, Resnick, E, Bradley, A.R, Collins, P, Brandao-Neto, J, Talon, R, Krojer, T, Bountra, C, Arrowsmith, C.H, Edwards, A, London, N, von Delft, F. | | 登録日 | 2018-08-10 | | 公開日 | 2019-12-18 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Covalent fragment group deposition
To Be Published
|
|
