7XLT
 
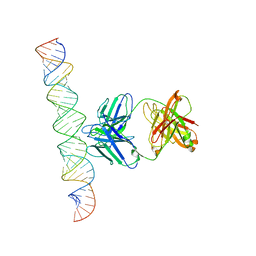 | | Cryo-EM Structure of R-loop monoclonal antibody S9.6 in recognizing RNA:DNA hybrids | | 分子名称: | DNA, RNA, S9.6 Fab HC, ... | | 著者 | Li, Q, Lin, C, Luo, Z, Li, H, Li, X, Sun, Q. | | 登録日 | 2022-04-22 | | 公開日 | 2022-05-25 | | 最終更新日 | 2022-08-03 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Cryo-EM structure of R-loop monoclonal antibody S9.6 in recognizing RNA:DNA hybrids.
J Genet Genomics, 49, 2022
|
|
3THZ
 
 | |
1OWL
 
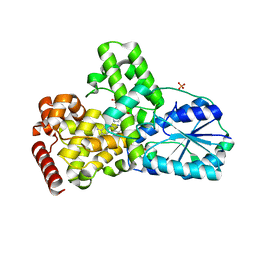 | | Structure of apophotolyase from Anacystis nidulans | | 分子名称: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | 著者 | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | 登録日 | 2003-03-28 | | 公開日 | 2004-04-13 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
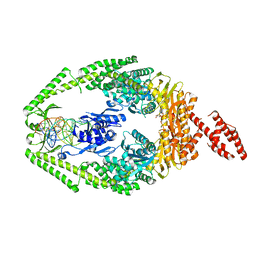
3THX
 
 | |
3THY
 
 | |
5I36
 
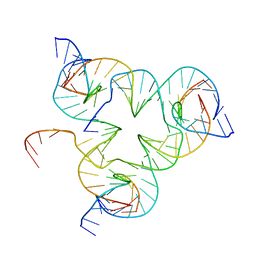 | | Crystal structure of color device state A | | 分子名称: | DNA (26-MER), DNA (5'-D(*AP*CP*AP*GP*TP*CP*GP*TP*GP*GP*TP*AP*TP*C)-3'), DNA (5'-D(*CP*AP*GP*AP*TP*AP*CP*CP*TP*GP*AP*TP*CP*GP*GP*AP*CP*TP*AP*CP*G)-3'), ... | | 著者 | Hao, Y, Kristiansen, M, Sha, R, Birktoft, J, Mao, C, Seeman, N.C. | | 登録日 | 2016-02-09 | | 公開日 | 2017-01-18 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (5.123 Å) | | 主引用文献 | A device that operates within a self-assembled 3D DNA crystal.
Nat Chem, 9, 2017
|
|
4B9Q
 
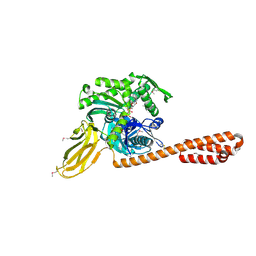 | |
8D93
 
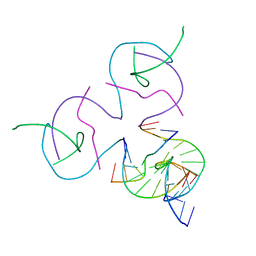 | | [2T7] Self-assembling tensegrity triangle with R3 symmetry at 2.96 A resolution, update and junction cut for entry 3GBI | | 分子名称: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*TP*GP*TP*GP*GP*CP*TP*GP*C)-3'), DNA (5'-D(P*AP*CP*AP*CP*CP*GP*T)-3'), ... | | 著者 | Vecchioni, S, Woloszyn, K, Lu, B, Sha, R, Ohayon, Y.P, Seeman, N.C. | | 登録日 | 2022-06-09 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.96 Å) | | 主引用文献 | The Rule of Thirds: Controlling Junction Chirality and Polarity in 3D DNA Tiles.
Small, 19, 2023
|
|
6FRE
 
 | | Crystal structure of G-1F/H73A mutant of Ssp DnaB Mini-Intein variant M86 | | 分子名称: | Replicative DNA helicase,Replicative DNA helicase | | 著者 | Popp, M.A, Blankenfeldt, W, Friedel, K, Mootz, H.D. | | 登録日 | 2018-02-15 | | 公開日 | 2019-01-30 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.22 Å) | | 主引用文献 | A functional interplay between intein and extein sequences in protein splicing compensates for the essential block B histidine.
Chem Sci, 10, 2019
|
|
6UAL
 
 | | A Self-Assembling DNA Crystal Scaffold with Cavities Containing 3 Helical Turns per Edge | | 分子名称: | DNA (32-MER), DNA (5'-D(*TP*GP*GP*AP*AP*AP*CP*AP*GP*AP*CP*TP*GP*TP*CP*AP*GP*AP*TP*G)-3'), DNA (5'-D(P*AP*GP*CP*AP*TP*GP*A)-3'), ... | | 著者 | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | 登録日 | 2019-09-10 | | 公開日 | 2020-09-16 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (4.514 Å) | | 主引用文献 | Crystal Structure of a Self-Assembling DNA Crystal Scaffold with Rhombohedral Symmetry
To Be Published
|
|
6FRH
 
 | | Crystal structure of Ssp DnaB Mini-Intein variant M86 | | 分子名称: | Replicative DNA helicase,Replicative DNA helicase | | 著者 | Popp, M.A, Blankenfeldt, W, Gazdag, M.E, Matern, J.J.C, Mootz, H.D. | | 登録日 | 2018-02-15 | | 公開日 | 2019-01-30 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | A functional interplay between intein and extein sequences in protein splicing compensates for the essential block B histidine.
Chem Sci, 10, 2019
|
|
1VTJ
 
 | | MOLECULAR STRUCTURE OF THE NETROPSIN-D(CGCGATATCGCG) COMPLEX: DNA CONFORMATION IN AN ALTERNATING AT SEGMENT; CONFORMATION 1 | | 分子名称: | DNA (5'-D(*CP*GP*CP*GP*AP*TP*AP*TP*CP*GP*CP*G)-3'), NETROPSIN | | 著者 | Coll, M, Aymami, J, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Wang, A.H.-J. | | 登録日 | 1999-09-14 | | 公開日 | 2011-07-13 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Molecular Structure of the Netropsin-d(CGCGATATCGCG) Complex: DNA Conformation in an Alternating AT Segment
Biochemistry, 28, 1989
|
|
7B4Z
 
 | | Synthetic DNA duplex dodecamer | | 分子名称: | DNA (5'-D(*CP*AP*CP*GP*CP*CP*GP*CP*TP*G)-3'), DNA (5'-D(*CP*AP*GP*CP*GP*GP*CP*GP*TP*G)-3') | | 著者 | Lomzov, A.A, Shernuykov, A.V, Sviridov, E.A, Shevelev, G.Y, Bagryanskaya, E.G, Pyshnyi, D.V. | | 登録日 | 2020-12-03 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Study of a DNA Duplex by Nuclear Magnetic Resonance and Molecular Dynamics Simulations. Validation of Pulsed Dipolar Electron Paramagnetic Resonance Distance Measurements Using Triarylmethyl-Based Spin Labels.
J Phys Chem B, 120, 2016
|
|
7ARE
 
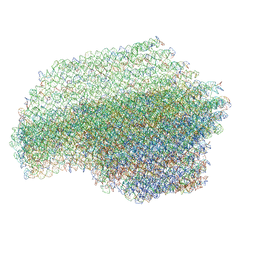 | | DNA origami pointer object v2 | | 分子名称: | SCAFFOLD STRAND, STAPLE STRAND | | 著者 | Thomas, M, Feigl, E, Kohler, F, Kube, M, Nagel-Yuksel, B, Willner, E.M, Funke, J.J, Gerling, T, Stommer, P, Honemann, M.N, Martin, T.G, Scheres, S.H.W, Dietz, H. | | 登録日 | 2020-10-24 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (7.4 Å) | | 主引用文献 | Revealing the structures of megadalton-scale DNA complexes with nucleotide resolution.
Nat Commun, 11, 2020
|
|
1S9B
 
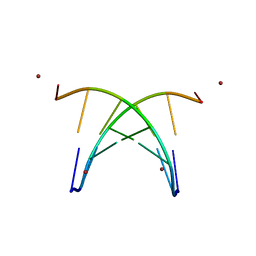 | | Crystal Structure Analysis of the B-DNA GAATTCG | | 分子名称: | 5'-D(*GP*AP*AP*TP*TP*CP*G)-3', NICKEL (II) ION | | 著者 | Valls, N, Uson, I, Gouyette, C, Subirana, J.A. | | 登録日 | 2004-02-04 | | 公開日 | 2004-09-07 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | A cubic arrangement of DNA double helices based on nickel-guanine interactions
J.Am.Chem.Soc., 126, 2004
|
|
7X4E
 
 | | Structure of 10635-DndE | | 分子名称: | DNA sulfur modification protein DndE, GLYCEROL | | 著者 | Haiyan, G, Wei, H, Chen, S, Wang, L, Wu, G. | | 登録日 | 2022-03-02 | | 公開日 | 2022-04-20 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Structural and Functional Analysis of DndE Involved in DNA Phosphorothioation in the Haloalkaliphilic Archaea Natronorubrum bangense JCM10635.
Mbio, 13, 2022
|
|
1VTC
 
 | |
6A7Y
 
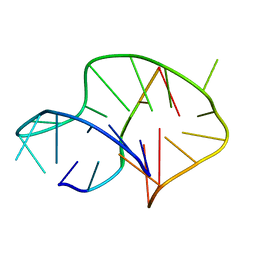 | | Solution structure of an intermolecular leaped V-shape G-quadruplex | | 分子名称: | DNA (5'-D(*GP*GP*TP*TP*TP*TP*GP*GP*GP*GP*TP*TP*TP*TP*GP*GP*GP*G)-3'), DNA (5'-D(*TP*GP*GP*GP*GP*A)-3') | | 著者 | Wan, C.J, Zhang, N. | | 登録日 | 2018-07-05 | | 公開日 | 2018-12-12 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR solution structure of an asymmetric intermolecular leaped V-shape G-quadruplex: selective recognition of the d(G2NG3NG4) sequence motif by a short linear G-rich DNA probe.
Nucleic Acids Res., 47, 2019
|
|
3HCL
 
 | | Helical superstructures in a DNA oligonucleotide crystal | | 分子名称: | DNA (5'-D(*CP*GP*AP*TP*AP*T)-3') | | 著者 | De Luchi, D, Martinez de Ilarduya, I, Subirana, J.A, Uson, I, Campos, J.L. | | 登録日 | 2009-05-06 | | 公開日 | 2010-05-19 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | A geometric approach to the crystallographic solution of nonconventional DNA structures: helical superstructures of d(CGATAT)
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
7N5K
 
 | | PCNA from Thermococcus gammatolerans: crystal II, collection 1, 1.98 A, 3.84 MGy | | 分子名称: | DNA polymerase sliding clamp, GLYCEROL, SULFATE ION | | 著者 | Marin-Tovar, Y, Rudino-Pinera, E. | | 登録日 | 2021-06-05 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | PCNA from Thermococcus gammatolerans: A protein involved in chromosomal DNA metabolism intrinsically resistant at high levels of ionizing radiation.
Proteins, 90, 2022
|
|
7N5M
 
 | | PCNA from Thermococcus gammatolerans: crystal III, collection 1, 2.00 A, 1.91 MGy | | 分子名称: | DNA polymerase sliding clamp, GLYCEROL, SULFATE ION | | 著者 | Marin-Tovar, Y, Rudino-Pinera, E. | | 登録日 | 2021-06-05 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | PCNA from Thermococcus gammatolerans: A protein involved in chromosomal DNA metabolism intrinsically resistant at high levels of ionizing radiation.
Proteins, 90, 2022
|
|
7N5J
 
 | | PCNA from Thermococcus gammatolerans: crystal I, collection 5, 2.82 A, 89.1 MGy | | 分子名称: | DNA polymerase sliding clamp, GLYCEROL, SULFATE ION | | 著者 | Marin-Tovar, Y, Rudino-Pinera, E. | | 登録日 | 2021-06-05 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.82 Å) | | 主引用文献 | PCNA from Thermococcus gammatolerans: A protein involved in chromosomal DNA metabolism intrinsically resistant at high levels of ionizing radiation.
Proteins, 90, 2022
|
|
7N5I
 
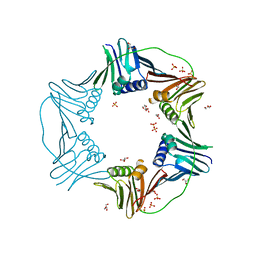 | | PCNA from Thermococcus gammatolerans: crystal I, collection 1, 1.95 A, 5.22 MGy | | 分子名称: | DNA polymerase sliding clamp, GLYCEROL, SULFATE ION | | 著者 | Marin-Tovar, Y, Rudino-Pinera, E. | | 登録日 | 2021-06-05 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | PCNA from Thermococcus gammatolerans: A protein involved in chromosomal DNA metabolism intrinsically resistant at high levels of ionizing radiation.
Proteins, 90, 2022
|
|
7N5L
 
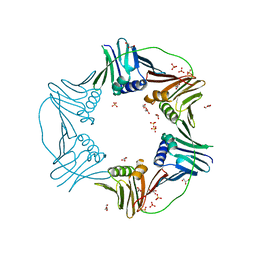 | | PCNA from Thermococcus gammatolerans: crystal II, collection 20, 3.07 A, 77.0 MGy | | 分子名称: | DNA polymerase sliding clamp, GLYCEROL, SULFATE ION | | 著者 | Marin-Tovar, Y, Rudino-Pinera, E. | | 登録日 | 2021-06-05 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.07 Å) | | 主引用文献 | PCNA from Thermococcus gammatolerans: A protein involved in chromosomal DNA metabolism intrinsically resistant at high levels of ionizing radiation.
Proteins, 90, 2022
|
|
7N5N
 
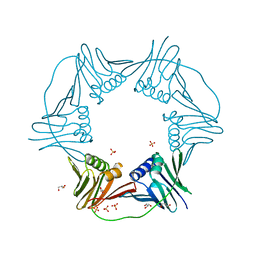 | | PCNA from Thermococcus gammatolerans: crystal III, collection 15, 2.20 A, 28.7 MGy | | 分子名称: | DNA polymerase sliding clamp, GLYCEROL, SULFATE ION | | 著者 | Marin-Tovar, Y, Rudino-Pinera, E. | | 登録日 | 2021-06-05 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | PCNA from Thermococcus gammatolerans: A protein involved in chromosomal DNA metabolism intrinsically resistant at high levels of ionizing radiation.
Proteins, 90, 2022
|
|
