1E3Z
 
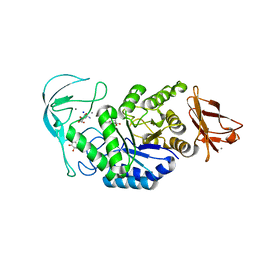 | | Acarbose complex of chimaeric amylase from B. amyloliquefaciens and B. licheniformis at 1.93A | | 分子名称: | 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, ... | | 著者 | Brzozowski, A.M, Lawson, D.M, Turkenburg, J.P, Bisgaard-Frantzen, H, Svendsen, A, Borchert, T.V, Dauter, Z, Wilson, K.S, Davies, G.J. | | 登録日 | 2000-06-27 | | 公開日 | 2001-06-21 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Structural Analysis of a Chimeric Bacterial Alpha-Amylase. High Resolution Analysis of Native and Ligand Complexes
Biochemistry, 39, 2000
|
|
1E40
 
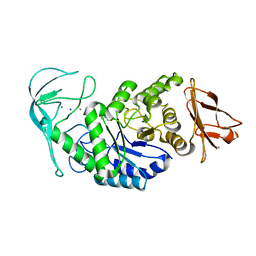 | | Tris/maltotriose complex of chimaeric amylase from B. amyloliquefaciens and B. licheniformis at 2.2A | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ALPHA-AMYLASE, CALCIUM ION, ... | | 著者 | Brzozowski, A.M, Lawson, D.M, Turkenburg, J.P, Bisgaard-Frantzen, H, Svendsen, A, Borchert, T.V, Dauter, Z, Wilson, K.S, Davies, G.J. | | 登録日 | 2000-06-27 | | 公開日 | 2001-06-21 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural Analysis of a Chimeric Bacterial Alpha-Amylase. High Resolution Analysis of Native and Ligand Complexes
Biochemistry, 39, 2000
|
|
1E41
 
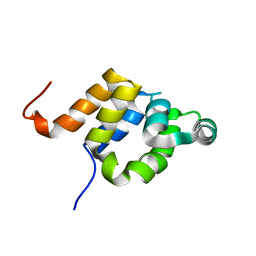 | |
1E42
 
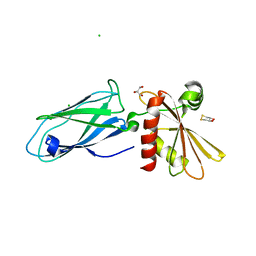 | | Beta2-adaptin appendage domain, from clathrin adaptor AP2 | | 分子名称: | AP-2 COMPLEX SUBUNIT BETA, CHLORIDE ION, DITHIANE DIOL, ... | | 著者 | Owen, D.J, Evans, P.R, McMahon, H.T. | | 登録日 | 2000-06-27 | | 公開日 | 2000-08-21 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The Structure and Function of the Beta2-Adaptin Appendage Domain
Embo J., 19, 2000
|
|
1E43
 
 | | Native structure of chimaeric amylase from B. amyloliquefaciens and B. licheniformis at 1.7A | | 分子名称: | ALPHA-AMYLASE, CALCIUM ION, SODIUM ION | | 著者 | Brzozowski, A.M, Lawson, D.M, Turkenburg, J.P, Bisgaard-Frantzen, H, Svendsen, A, Borchert, T.V, Dauter, Z, Wilson, K.S, Davies, G.J. | | 登録日 | 2000-06-27 | | 公開日 | 2001-06-21 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural Analysis of a Chimeric Bacterial Alpha-Amylase. High Resolution Analysis of Native and Ligand Complexes
Biochemistry, 39, 2000
|
|
1E44
 
 | | ribonuclease domain of colicin E3 in complex with its immunity protein | | 分子名称: | 1,2-ETHANEDIOL, COLICIN E3, IMMUNITY PROTEIN | | 著者 | Carr, S, Walker, D, James, R, Kleanthous, C, Hemmings, A.M. | | 登録日 | 2000-06-28 | | 公開日 | 2001-06-28 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Inhibition of a Ribosome Inactivating Ribonuclease: The Crystal Structure of the Cytotoxic Domain of Colicin E3 in Complex with its Immunity Protein
Structure, 8, 2000
|
|
1E46
 
 | |
1E47
 
 | |
1E48
 
 | |
1E49
 
 | |
1E4A
 
 | |
1E4B
 
 | |
1E4C
 
 | |
1E4D
 
 | | Structure of OXA10 beta-lactamase at pH 8.3 | | 分子名称: | 1,2-ETHANEDIOL, BETA-LACTAMASE OXA-10, SULFATE ION | | 著者 | Maveyraud, L, Golemi, D, Kotra, L.P, Tranier, S, Vakulenko, S, Mobashery, S, Samama, J.P. | | 登録日 | 2000-07-03 | | 公開日 | 2001-01-12 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Insights Into Class D Beta-Lactamases are Revealed by the Crystal Structure of the Oxa10 Enzyme from Pseudomonas Aeruginosa
Structure, 8, 2000
|
|
1E4E
 
 | | D-alanyl-D-lacate ligase | | 分子名称: | 1(S)-AMINOETHYL-(2-CARBOXYPROPYL)PHOSPHORYL-PHOSPHINIC ACID, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | 著者 | Roper, D.I. | | 登録日 | 2000-07-03 | | 公開日 | 2001-06-28 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The molecular basis of vancomycin resistance in clinically relevant Enterococci: crystal structure of D-alanyl-D-lactate ligase (VanA).
Proc. Natl. Acad. Sci. U.S.A., 97, 2000
|
|
1E4F
 
 | |
1E4G
 
 | |
1E4H
 
 | | Structure of human transthyretin complexed with bromophenols: a new mode of binding | | 分子名称: | GLYCEROL, PENTABROMOPHENOL, TRANSTHYRETIN | | 著者 | Ghosh, M, Meerts, I.A.T.M, Cook, A, Bergman, A, Brouwer, A, Johnson, L.N. | | 登録日 | 2000-07-04 | | 公開日 | 2000-08-29 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of Human Transthyretin Complexed with Bromophenols : A New Mode of Binding
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1E4I
 
 | | 2-deoxy-2-fluoro-beta-D-glucosyl/enzyme intermediate complex of the beta-glucosidase from Bacillus polymyxa | | 分子名称: | 2,4-dinitrophenyl 2-deoxy-2-fluoro-beta-D-glucopyranoside, 2-deoxy-2-fluoro-alpha-D-glucopyranose, BETA-GLUCOSIDASE | | 著者 | Sanz-Aparicio, J, Gonzalez, B, Hermoso, J.A, Arribas, J.C, Canada, F.J, Polaina, J. | | 登録日 | 2000-07-06 | | 公開日 | 2001-07-05 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Basis of Increased Resistance to Thermal Denaturation Induced by Single Amino Acid Substitution in the Sequence of Beta-Glucosidase a from Bacillus Polymyxa.
Proteins: Struct.,Funct., Genet., 33, 1998
|
|
1E4J
 
 | |
1E4K
 
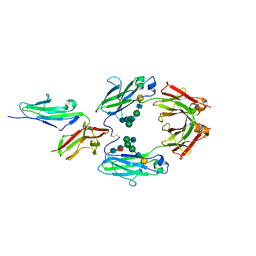 | | CRYSTAL STRUCTURE OF SOLUBLE HUMAN IGG1 FC FRAGMENT-FC-GAMMA RECEPTOR III COMPLEX | | 分子名称: | FC FRAGMENT OF HUMAN IGG1, LOW AFFINITY IMMUNOGLOBULIN GAMMA FC RECEPTOR III, alpha-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Sondermann, P, Huber, R, Oosthuizen, V, Jacob, U. | | 登録日 | 2000-07-07 | | 公開日 | 2000-08-06 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | The 3.2A Crystal Structure of the Human Igg1 Fc Fragment-Fc-Gamma-Riii Complex
Nature, 406, 2000
|
|
1E4L
 
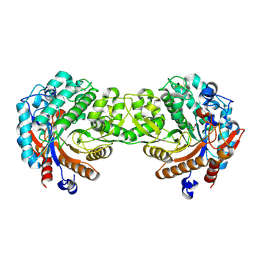 | | Crystal structure of the inactive mutant Monocot (Maize ZMGlu1) beta-glucosidase ZM Glu191Asp | | 分子名称: | BETA-GLUCOSIDASE, CHLOROPLASTIC, GLYCEROL | | 著者 | Czjzek, M, Cicek, M, Bevan, D.R, Henrissat, B, Esen, A. | | 登録日 | 2000-07-10 | | 公開日 | 2000-12-11 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The Mechanism of Substrate (Aglycone) Specificity in Beta -Glucosidases is Revealed by Crystal Structures of Mutant Maize Beta -Glucosidase-Dimboa, -Dimboaglc, and -Dhurrin Complexes
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1E4M
 
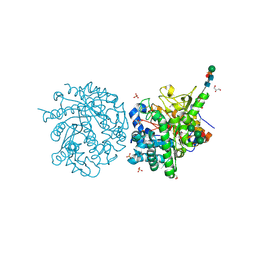 | | MYROSINASE FROM SINAPIS ALBA | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | 著者 | Burmeister, W.P. | | 登録日 | 2000-07-10 | | 公開日 | 2001-05-25 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | High Resolution X-Ray Crystallography Shows that Ascorbate is a Cofactor for Myrosinase and Substitutes for the Function of the Catalytic Base
J.Biol.Chem., 275, 2000
|
|
1E4N
 
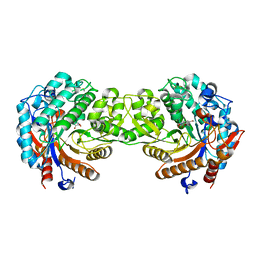 | | Crystal structure of the inactive mutant Monocot (Maize ZMGlu1) beta-glucosidase ZMGluE191D in complex with the natural aglycone DIMBOA | | 分子名称: | 2,4-DIHYDROXY-7-(METHYLOXY)-2H-1,4-BENZOXAZIN-3(4H)-ONE, BETA-GLUCOSIDASE | | 著者 | Czjzek, M, Cicek, M, Bevan, D.R, Zamboni, V, Henrissat, B, Esen, A. | | 登録日 | 2000-07-11 | | 公開日 | 2000-12-11 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The Mechanism of Substrate (Aglycone) Specificity in Beta -Glucosidases is Revealed by Crystal Structures of Mutant Maize Beta -Glucosidase-Dimboa, -Dimboaglc, and -Dhurrin Complexes
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1E4O
 
 | | Phosphorylase recognition and phosphorolysis of its oligosaccharide substrate: answers to a long outstanding question | | 分子名称: | MALTODEXTRIN PHOSPHORYLASE, PYRIDOXAL-5'-PHOSPHATE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Watson, K.A, McCleverty, C, Geremia, S, Cottaz, S, Driguez, H, Johnson, L.N. | | 登録日 | 2000-07-11 | | 公開日 | 2000-08-16 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Phosphorylase Recognition and Phosphorylysis of its Oligosaccharide Substrate: Answers to a Long Outstanding Question
Embo J., 18, 1999
|
|
