2Z1M
 
 | |
2Z5U
 
 | |
3BX7
 
 | |
3BYI
 
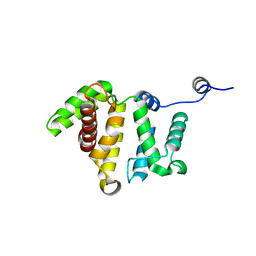 | | Crystal structure of human Rho GTPase activating protein 15 (ARHGAP15) | | 分子名称: | Rho GTPase activating protein 15 | | 著者 | Shrestha, L, Tickle, J, Elkins, J, Burgess-Brown, N, Johansson, C, Papagrigoriou, E, Kavanagh, K, Pike, A.C.W, Ugochukwu, E, Uppenberg, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Doyle, D, Structural Genomics Consortium (SGC) | | 登録日 | 2008-01-16 | | 公開日 | 2008-02-26 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Crystal Structure of Human Rho GTPase Activating Protein 15 (ARHGAP15).
To be Published
|
|
3BKU
 
 | |
3C7Z
 
 | |
3BQW
 
 | |
3BKQ
 
 | |
3ARO
 
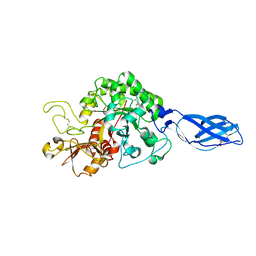 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - apo structure | | 分子名称: | Chitinase A, GLYCEROL | | 著者 | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | 登録日 | 2010-12-09 | | 公開日 | 2011-04-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
3ARY
 
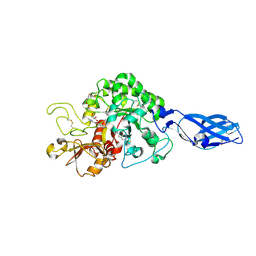 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - complex structure with 2-(imidazolin-2-yl)-5-isothiocyanatobenzofuran | | 分子名称: | 2-(5-isothiocyanato-1-benzofuran-2-yl)-4,5-dihydro-1H-imidazole, Chitinase A, GLYCEROL | | 著者 | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | 登録日 | 2010-12-09 | | 公開日 | 2011-04-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
3ARX
 
 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - complex structure with Propentofylline | | 分子名称: | 3-methyl-1-(5-oxohexyl)-7-propyl-3,7-dihydro-1H-purine-2,6-dione, Chitinase A, GLYCEROL | | 著者 | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | 登録日 | 2010-12-09 | | 公開日 | 2011-04-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.16 Å) | | 主引用文献 | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
3ASU
 
 | |
3ARP
 
 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - complex structure with DEQUALINIUM | | 分子名称: | Chitinase A, DEQUALINIUM, GLYCEROL | | 著者 | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | 登録日 | 2010-12-09 | | 公開日 | 2011-04-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
3ARW
 
 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - complex structure with chelerythrine | | 分子名称: | 1,2-dimethoxy-12-methyl[1,3]benzodioxolo[5,6-c]phenanthridin-12-ium, Chitinase A, GLYCEROL | | 著者 | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | 登録日 | 2010-12-09 | | 公開日 | 2011-04-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
3C81
 
 | |
3C8S
 
 | |
3C7C
 
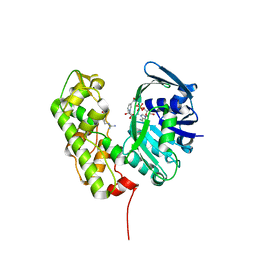 | | A structural basis for substrate and stereo selectivity in octopine dehydrogenase (ODH-NADH-L-Arginine) | | 分子名称: | ARGININE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Octopine dehydrogenase | | 著者 | Smits, S.H.J, Mueller, A, Schmitt, L, Grieshaber, M.K. | | 登録日 | 2008-02-07 | | 公開日 | 2008-07-22 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | A structural basis for substrate selectivity and stereoselectivity in octopine dehydrogenase from Pecten maximus.
J.Mol.Biol., 381, 2008
|
|
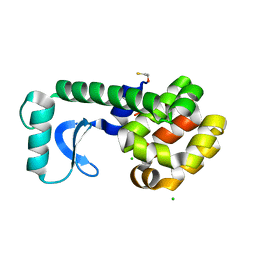
3C83
 
 | |
3BXV
 
 | |
3BUO
 
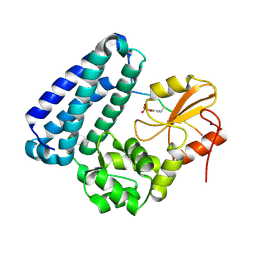 | | Crystal structure of c-Cbl-TKB domain complexed with its binding motif in EGF receptor' | | 分子名称: | 13-meric peptide from Epidermal growth factor receptor, E3 ubiquitin-protein ligase CBL | | 著者 | Ng, C, Jackson, R.A, Buschdorf, J.P, Sun, Q, Guy, G.R, Sivaraman, J. | | 登録日 | 2008-01-03 | | 公開日 | 2008-02-26 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis for a novel intrapeptidyl H-bond and reverse binding of c-Cbl-TKB domain substrates
Embo J., 27, 2008
|
|
3AAR
 
 | | Crystal structure of Lp1NTPDase from Legionella pneumophila in complex with AMPPNP | | 分子名称: | Ectonucleoside triphosphate diphosphohydrolase I, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Ge, H, Vivian, J.P, Beddoe, T, Rossjohn, J. | | 登録日 | 2009-11-24 | | 公開日 | 2010-02-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal Structure of a Legionella pneumophila Ecto -Triphosphate Diphosphohydrolase, A Structural and Functional Homolog of the Eukaryotic NTPDases
Structure, 18, 2010
|
|
3AB9
 
 | | Crystal Structure of lipoylated E. coli H-protein (reduced form) | | 分子名称: | CALCIUM ION, CHLORIDE ION, Glycine cleavage system H protein | | 著者 | Okamura-Ikeda, K, Maita, N. | | 登録日 | 2009-12-04 | | 公開日 | 2010-04-07 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structure of aminomethyltransferase in complex with dihydrolipoyl-H-protein of the glycine cleavage system: implications for recognition of lipoyl protein substrate, disease-related mutations, and reaction mechanism
J.Biol.Chem., 285, 2010
|
|
3A5Y
 
 | | Crystal structure of GenX from Escherichia coli in complex with lysyladenylate analog | | 分子名称: | 5'-O-[(L-LYSYLAMINO)SULFONYL]ADENOSINE, Putative lysyl-tRNA synthetase | | 著者 | Sumida, T, Yanagisawa, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2009-08-17 | | 公開日 | 2010-08-25 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A paralog of lysyl-tRNA synthetase aminoacylates a conserved lysine residue in translation elongation factor P.
Nat.Struct.Mol.Biol., 17, 2010
|
|
336D
 
 | | INTERACTION BETWEEN LEFT-HANDED Z-DNA AND POLYAMINE-3 THE CRYSTAL STRUCTURE OF THE D(CG)3 AND THERMOSPERMINE COMPLEX | | 分子名称: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), MAGNESIUM ION, N-(3-AMINO-PROPYL)-N-(5-AMINOPROPYL)-1,4-DIAMINOBUTANE | | 著者 | Ohishi, H, Terasoma, N, Nakanishi, I, Van Der Marel, G, Van Boom, J.H, Rich, A, Wang, A.H.-J, Hakoshima, T, Tomita, K.-I. | | 登録日 | 1997-06-24 | | 公開日 | 1998-04-10 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Interaction between left-handed Z-DNA and polyamine - 3. The crystal structure of the d(CG)3 and thermospermine complex.
FEBS Lett., 398, 1996
|
|
3ACF
 
 | | Crystal Structure of Carbohydrate-Binding Module Family 28 from Clostridium josui Cel5A in a ligand-free form | | 分子名称: | Beta-1,4-endoglucanase, CALCIUM ION, SULFATE ION | | 著者 | Tsukimoto, K, Takada, R, Araki, Y, Suzuki, K, Karita, S, Wakagi, T, Shoun, H, Watanabe, T, Fushinobu, S. | | 登録日 | 2010-01-04 | | 公開日 | 2010-03-02 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Recognition of cellooligosaccharides by a family 28 carbohydrate-binding module.
Febs Lett., 584, 2010
|
|
