3OAN
 
 | | Crystal structure of the Ran Binding Domain From The Nuclear Complex Component Nup2 From Ashbya Gossypii | | 分子名称: | ABR034Wp, GLYCEROL | | 著者 | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2010-08-05 | | 公開日 | 2010-08-18 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of the Ran Binding Domain From The
Nuclear Complex Component Nup2 From Ashbya Gossypii
To be Published
|
|
3WSO
 
 | | Crystal structure of the Skp1-FBG3 complex | | 分子名称: | F-box only protein 44, S-phase kinase-associated protein 1 | | 著者 | Kumanomidou, T, Nishio, K, Takagi, K, Nakagawa, T, Suzuki, A, Yamane, T, Tokunaga, F, Iwai, K, Murakami, A, Yoshida, Y, Tanaka, K, Mizushima, T. | | 登録日 | 2014-03-18 | | 公開日 | 2015-03-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The Structural Differences between a Glycoprotein Specific F-Box Protein Fbs1 and Its Homologous Protein FBG3
Plos One, 10, 2015
|
|
3NOJ
 
 | | The structure of HMG/CHA aldolase from the protocatechuate degradation pathway of Pseudomonas putida | | 分子名称: | 4-carboxy-4-hydroxy-2-oxoadipate aldolase/oxaloacetate decarboxylase, MAGNESIUM ION, PYRUVIC ACID, ... | | 著者 | Kimber, M.S, Wang, W, Mazurkewich, S, Seah, S.Y.K. | | 登録日 | 2010-06-25 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Structural and Kinetic Characterization of 4-Hydroxy-4-methyl-2-oxoglutarate/4-Carboxy-4-hydroxy-2-oxoadipate Aldolase, a Protocatechuate Degradation Enzyme Evolutionarily Convergent with the HpaI and DmpG Pyruvate Aldolases.
J.Biol.Chem., 285, 2010
|
|
3O4L
 
 | | Genetic and structural basis for selection of a ubiquitous T cell receptor deployed in Epstein-Barr virus | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BSLF2/BMLF1 protein, Beta-2-microglobulin, ... | | 著者 | Miles, J.J, Bulek, A.M, Cole, D.K, Gostick, E, Schauenburg, J.A, Dolton, G, Venturi, V, Davenport, M.P, Tan, M.P, Burrows, S.R, Wooldridge, L, Price, D.A, Rizkallah, P.J, Sewell, A.K. | | 登録日 | 2010-07-27 | | 公開日 | 2011-01-12 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | Genetic and structural basis for selection of a ubiquitous T cell receptor deployed in Epstein-Barr virus infection.
Plos Pathog., 6, 2010
|
|
3WIZ
 
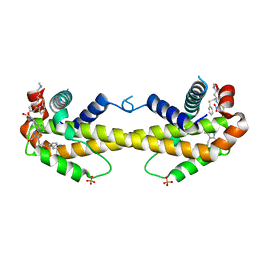 | | Crystal structure of Bcl-xL in complex with compound 10 | | 分子名称: | 7-(4-{[(4-{[(2R)-4-(dimethylamino)-1-(phenylsulfanyl)butan-2-yl]amino}-3-nitrophenyl)sulfonyl]carbamoyl}-2-methylphenyl)-3-[3-(naphthalen-1-yloxy)propyl]pyrazolo[1,5-a]pyridine-2-carboxylic acid, Bcl-2-like protein 1, PHOSPHATE ION | | 著者 | Sogabe, S, Igaki, S, Hayano, Y. | | 登録日 | 2013-09-26 | | 公開日 | 2013-11-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Discovery of potent Mcl-1/Bcl-xL dual inhibitors by using a hybridization strategy based on structural analysis of target proteins.
J.Med.Chem., 56, 2013
|
|
3WXS
 
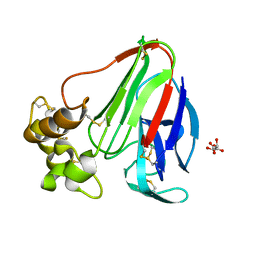 | | Thaumatin structure determined by SPring-8 Angstrom Compact free electron Laser (SACLA) | | 分子名称: | L(+)-TARTARIC ACID, thaumatin I | | 著者 | Masuda, T, Nango, E, Sugahara, M, Mizohata, E, Tanaka, T, Tanaka, R, Suzuki, M, Mikami, B, Iwata, S. | | 登録日 | 2014-08-07 | | 公開日 | 2014-11-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Grease matrix as a versatile carrier of proteins for serial crystallography
Nat. Methods, 12, 2015
|
|
3O7A
 
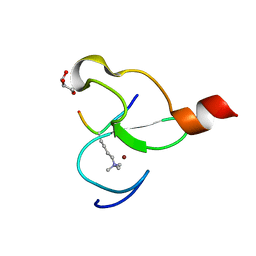 | | Crystal structure of PHF13 in complex with H3K4me3 | | 分子名称: | GLYCEROL, H3K4ME3 HISTONE 11MER-PEPTIDE, PHD finger protein 13 variant, ... | | 著者 | Bian, C.B, Lam, R, Xu, C, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2010-07-30 | | 公開日 | 2010-10-06 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | PHF13 is a molecular reader and transcriptional co-regulator of H3K4me2/3.
Elife, 5, 2016
|
|
3OB6
 
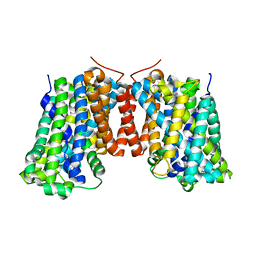 | | Structure of AdiC(N101A) in the open-to-out Arg+ bound conformation | | 分子名称: | ARGININE, AdiC arginine:agmatine antiporter | | 著者 | Carpena, X, Kowalczyk, L, Ratera, M, Valencia, E, Vazquez-lbar, J.L, Fita, I, Palacin, M. | | 登録日 | 2010-08-06 | | 公開日 | 2011-03-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Molecular basis of substrate-induced permeation by an amino acid antiporter.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3NS5
 
 | |
4A7C
 
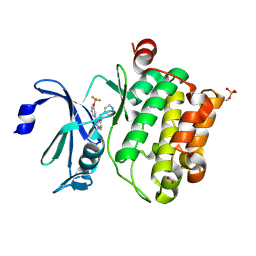 | | Crystal structure of PIM1 kinase with ETP46546 | | 分子名称: | ACETATE ION, IMIDAZOLE, N-(piperidin-4-ylmethyl)-3-[3-(trifluoromethyloxy)phenyl]-[1,2,3]triazolo[4,5-b]pyridin-5-amine, ... | | 著者 | Mazzorana, M, Montoya, G. | | 登録日 | 2011-11-12 | | 公開日 | 2012-02-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Hit to Lead Evaluation of 1,2,3-Triazolo[4,5-B]Pyridines as Pim Kinase Inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4A1X
 
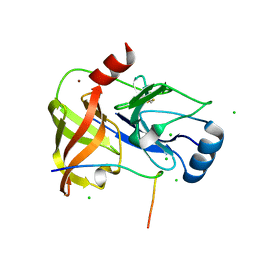 | | Co-Complex structure of NS3-4A protease with the inhibitory peptide CP5-46-A (Synchrotron data) | | 分子名称: | CHLORIDE ION, CP5-46-A PEPTIDE, NONSTRUCTURAL PROTEIN 4A, ... | | 著者 | Schmelz, S, Kuegler, J, Collins, J, Heinz, D. | | 登録日 | 2011-09-20 | | 公開日 | 2012-09-19 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | High Affinity Peptide Inhibitors of the Hepatitis C Virus Ns3-4A Protease Refractory to Common Resistant Mutants.
J.Biol.Chem., 287, 2012
|
|
4AF1
 
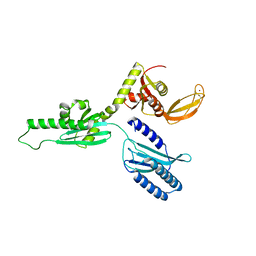 | |
3NSS
 
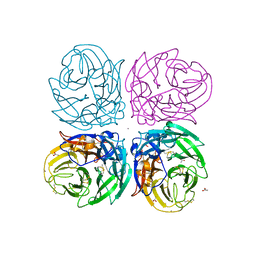 | | The 2009 pandemic H1N1 neuraminidase N1 lacks the 150-cavity in its active sites | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | 著者 | Li, Q, Qi, J.X, Zhang, W, Vavricka, C.J, Shi, Y, Gao, G.F. | | 登録日 | 2010-07-02 | | 公開日 | 2010-09-22 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.902 Å) | | 主引用文献 | The 2009 pandemic H1N1 neuraminidase N1 lacks the 150-cavity in its active site
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NU1
 
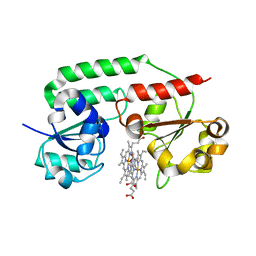 | | Structure of holo form of a periplasmic heme binding protein | | 分子名称: | Hemin-binding periplasmic protein, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Mattle, D, Goetz, B.A, Woo, J.S, Locher, K.P. | | 登録日 | 2010-07-06 | | 公開日 | 2010-10-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Two stacked heme molecules in the binding pocket of the periplasmic heme-binding protein HmuT from Yersinia pestis.
J.Mol.Biol., 404, 2010
|
|
4ALI
 
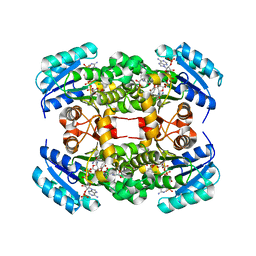 | | Crystal structure of S. aureus FabI in complex with NADP and triclosan (P1) | | 分子名称: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Schiebel, J, Chang, A, Tonge, P.J, Kisker, C. | | 登録日 | 2012-03-04 | | 公開日 | 2012-05-09 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Staphylococcus Aureus Fabi: Inhibition, Substrate Recognition and Potential Implications for in Vivo Essentiality
Structure, 20, 2012
|
|
4ADS
 
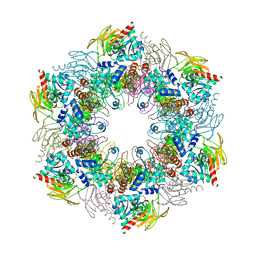 | | Crystal structure of plasmodial PLP synthase complex | | 分子名称: | PDX2 PROTEIN, PHOSPHATE ION, PYRIDOXINE BIOSYNTHETIC ENZYME PDX1 HOMOLOGUE, ... | | 著者 | Guedez, G, Sinning, I, Tews, I. | | 登録日 | 2012-01-03 | | 公開日 | 2012-01-25 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.61 Å) | | 主引用文献 | Assembly of the Eukaryotic Plp-Synthase Complex from Plasmodium and Activation of the Pdx1 Enzyme.
Structure, 20, 2012
|
|
3OHH
 
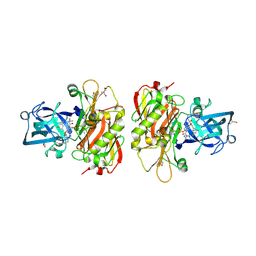 | | Crystal structure of beta-site app-cleaving enzyme 1 (bace-wt) complex with bms-681889 aka n~1~-butyl-5-cyano- n~3~-((1s,2r)-1-(3,5-difluorobenzyl)-2-hydroxy-3-((3- methoxybenzyl)amino)propyl)-n~1~-methyl-1h-indole-1,3- dicarboxamide | | 分子名称: | Beta-secretase 1, GLYCEROL, N~1~-butyl-5-cyano-N~3~-{(1S,2R)-1-(3,5-difluorobenzyl)-2-hydroxy-3-[(3-methoxybenzyl)amino]propyl}-N~1~-methyl-1H-indole-1,3-dicarboxamide, ... | | 著者 | Muckelbauer, J.K. | | 登録日 | 2010-08-17 | | 公開日 | 2011-04-06 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Synthesis and SAR of indole-and 7-azaindole-1,3-dicarboxamide hydroxyethylamine inhibitors of BACE-1.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4AL6
 
 | |
4A1G
 
 | | The crystal structure of the human Bub1 TPR domain in complex with the KI motif of Knl1 | | 分子名称: | MITOTIC CHECKPOINT SERINE/THREONINE-PROTEIN KINASE BUB1, PROTEIN CASC5 | | 著者 | Krenn, V, Wehenkel, A, Li, X, Santaguida, S, Musacchio, A. | | 登録日 | 2011-09-15 | | 公開日 | 2012-02-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural Analysis Reveals Features of the Spindle Checkpoint Kinase Bub1-Kinetochore Subunit Knl1 Interaction.
J.Cell Biol., 196, 2012
|
|
4A1V
 
 | | Co-Complex structure of NS3-4A protease with the optimized inhibitory peptide CP5-46A-4D5E | | 分子名称: | CHLORIDE ION, CP5-46A-4D5E, NON-STRUCTURAL PROTEIN 4A, ... | | 著者 | Schmelz, S, Kuegler, J, Collins, J, Heinz, D.W. | | 登録日 | 2011-09-20 | | 公開日 | 2012-09-19 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | High Affinity Peptide Inhibitors of the Hepatitis C Virus Ns3-4A Protease Refractory to Common Resistant Mutants.
J.Biol.Chem., 287, 2012
|
|
4AJT
 
 | |
3OFV
 
 | | Crystal structure of peptidyl-tRNA hydrolase from Escherichia Coli, I222 crystal form | | 分子名称: | Peptidyl-tRNA hydrolase | | 著者 | Lam, R, McGrath, T.E, Romanov, V, Gothe, S.A, Peddi, S.R, Razumova, E, Lipman, R.S, Branstrom, A.A, Chirgadze, N.Y. | | 登録日 | 2010-08-16 | | 公開日 | 2011-08-17 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Crystal structure of peptidyl-tRNA hydrolase from Escherichia Coli, I222 crystal form
To be Published
|
|
4AOH
 
 | | Structural snapshots and functional analysis of human angiogenin variants associated with Amyotrophic Lateral Sclerosis (ALS) | | 分子名称: | ANGIOGENIN, D(-)-TARTARIC ACID, L(+)-TARTARIC ACID | | 著者 | Thiyagarajan, N, Ferguson, R, Subramanian, V, Acharya, K.R. | | 登録日 | 2012-03-27 | | 公開日 | 2012-10-10 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.041 Å) | | 主引用文献 | Structural and Molecular Insights Into the Mechanism of Action of Human Angiogenin-Als Variants in Neurons
Nat.Commun., 3, 2012
|
|
3OHF
 
 | | Crystal structure of beta-site app-cleaving enzyme 1 (BACE-WT) complex with bms-655295 aka n~3~-((1s,2r)-1- benzyl-2-hydroxy-3-((3-methoxybenzyl)amino)propyl)-n~1~, n~1~-dibutyl-1h-indole-1,3-dicarboxamide | | 分子名称: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | 著者 | Muckelbauer, J.K. | | 登録日 | 2010-08-17 | | 公開日 | 2011-04-06 | | 最終更新日 | 2017-03-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Synthesis and SAR of indole-and 7-azaindole-1,3-dicarboxamide hydroxyethylamine inhibitors of BACE-1.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4AL3
 
 | |
