6S5X
 
 | | Structure of RibR, the most N-terminal Rib domain from Group B Streptococcus species Streptococcus agalactiae | | 分子名称: | Group B streptococcal R4 surface protein, SODIUM ION | | 著者 | Whelan, F, Turkenburg, J.P, Griffiths, S.C, Bateman, A, Potts, J.R. | | 登録日 | 2019-07-02 | | 公開日 | 2019-12-11 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Defining the remarkable structural malleability of a bacterial surface protein Rib domain implicated in infection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7YPY
 
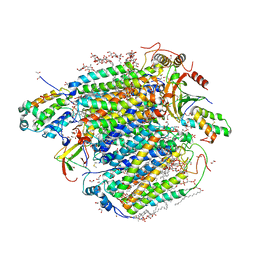 | | Bovine heart cytochrome c oxidase in fully oxidized state at 1.5 angstrom resolution | | 分子名称: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | 著者 | Shimada, A, Tsukihara, T. | | 登録日 | 2022-08-05 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The Mg2+-containing water cluster of mammalian cytochrome c oxidase collects four pumping proton equivalents in each catalytic cycle.
J. Biol. Chem., 291, 2016
|
|
6RP6
 
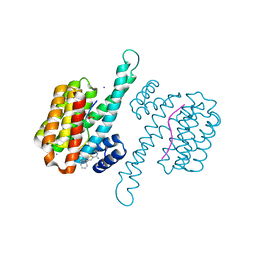 | | Fragment AZ-019 binding at the TAZpS89/14-3-3 sigma interface | | 分子名称: | 14-3-3 protein sigma, 4-phenyl-5-(piperidin-4-ylmethyl)thiophene-2-carboximidamide, CALCIUM ION, ... | | 著者 | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Patel, J, Castaldi, P, Ottmann, C. | | 登録日 | 2019-05-14 | | 公開日 | 2020-06-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.885 Å) | | 主引用文献 | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
4KLQ
 
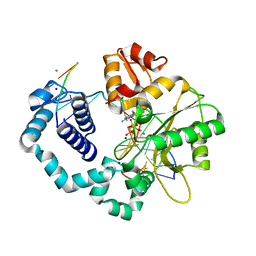 | | Observing a DNA polymerase choose right from wrong. | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*A)-3', ... | | 著者 | Freudenthal, B.D, Beard, W.A, Shock, D.D, Wilson, S.H. | | 登録日 | 2013-05-07 | | 公開日 | 2013-07-17 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.999 Å) | | 主引用文献 | Observing a DNA polymerase choose right from wrong.
Cell(Cambridge,Mass.), 154, 2013
|
|
7Z85
 
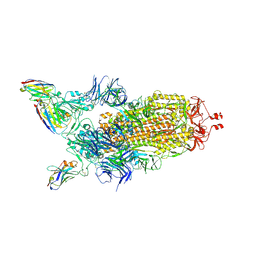 | | CRYO-EM STRUCTURE OF SARS-COV-2 SPIKE : H11-B5 nanobody complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody H11-B5, ... | | 著者 | Weckener, M, Naismith, J.H. | | 登録日 | 2022-03-16 | | 公開日 | 2022-07-13 | | 最終更新日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z9Q
 
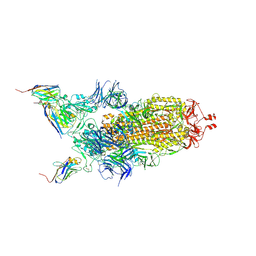 | | CRYO-EM STRUCTURE OF SARS-COV-2 SPIKE : H11-A10 nanobody complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody H11-A10, ... | | 著者 | Weckener, M, Naismith, J.H. | | 登録日 | 2022-03-21 | | 公開日 | 2022-07-13 | | 最終更新日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4N7S
 
 | | Crystal structure of Tse3-Tsi3 complex with Zinc ion | | 分子名称: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Shang, G.J. | | 登録日 | 2013-10-16 | | 公開日 | 2014-04-23 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.101 Å) | | 主引用文献 | Structural insights into the T6SS effector protein Tse3 and the Tse3-Tsi3 complex from Pseudomonas aeruginosa reveal a calcium-dependent membrane-binding mechanism
Mol.Microbiol., 92, 2014
|
|
7Z7X
 
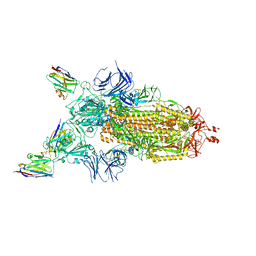 | | CRYO-EM STRUCTURE OF SARS-COV-2 SPIKE : H11-H6 nanobody complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody H11-H6, ... | | 著者 | Weckener, M, Naismith, J.H. | | 登録日 | 2022-03-16 | | 公開日 | 2022-07-13 | | 最終更新日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z86
 
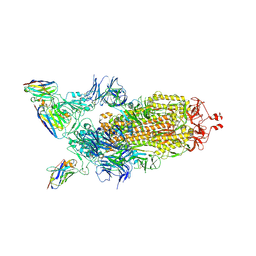 | |
7Z9R
 
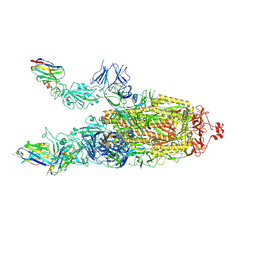 | |
5XC1
 
 | | Crystal structure of the complex of an aromatic mutant (W6A) of an alkali thermostable GH10 Xylanase from Bacillus sp. NG-27 with S-1,2-Propanediol | | 分子名称: | Beta-xylanase, MAGNESIUM ION, S-1,2-PROPANEDIOL, ... | | 著者 | Bansia, H, Mahanta, P, Ramakumar, S. | | 登録日 | 2017-03-21 | | 公開日 | 2018-03-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Small Glycols Discover Cryptic Pockets on Proteins for Fragment-Based Approaches.
J.Chem.Inf.Model., 2021
|
|
7Z6V
 
 | | CRYO-EM STRUCTURE OF SARS-COV-2 SPIKE : H11 nanobody complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody H11, ... | | 著者 | Weckener, M, Naismith, J.H, Vogirala, V.K. | | 登録日 | 2022-03-14 | | 公開日 | 2022-07-13 | | 最終更新日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4N3V
 
 | |
5EOZ
 
 | | Mutagenicity of 7-Benzyl guanine lesion and Replication by Human DNA Polymerase beta | | 分子名称: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*CP*CP*GP*AP*CP*(GFL)P*TP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*A)-3'), ... | | 著者 | Koag, M.C, Lee, S. | | 登録日 | 2015-11-10 | | 公開日 | 2016-11-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.088 Å) | | 主引用文献 | Structural and kinetic studies of the effect of guanine-N7 alkylation and metal cofactors on DNA replication.
Biochemistry, 2018
|
|
6CEK
 
 | |
5XVD
 
 | | [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 in an air-oxidized condition | | 分子名称: | FE3-S4 CLUSTER, FE4-S4-O CLUSTER, GLYCEROL, ... | | 著者 | Nishikawa, K, Matsuura, H, Muhd Noor, N.D, Tai, H, Hirota, S, Kim, J, Kang, J, Tateno, M, Yoon, K.S, Ogo, S, Shomura, Y, Higuchi, Y. | | 登録日 | 2017-06-27 | | 公開日 | 2018-06-27 | | 最終更新日 | 2019-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Chem.Commun.(Camb.), 54, 2018
|
|
6C9R
 
 | | Mycobacterium tuberculosis adenosine kinase bound to (2R,3S,4R,5R)-2-(hydroxymethyl)-5-(6-(thiophen-3-yl)-9H-purin-9-yl)tetrahydrofuran-3,4-diol | | 分子名称: | 9-beta-D-ribofuranosyl-6-(thiophen-3-yl)-9H-purine, Adenosine kinase, GLYCEROL, ... | | 著者 | Crespo, R.A, TB Structural Genomics Consortium (TBSGC) | | 登録日 | 2018-01-28 | | 公開日 | 2019-05-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure-Guided Drug Design of 6-Substituted Adenosine Analogues as Potent Inhibitors of Mycobacterium tuberculosis Adenosine Kinase.
J.Med.Chem., 62, 2019
|
|
6BZ2
 
 | | Crystal structure of wild-type HIV-1 protease with a novel HIV-1 inhibitor GRL-14213A of 6-5-5-ring fused crown-like tetrahydropyranofuran as the P2-ligand, a cyclopropylaminobenzothiazole as the P2'-ligand and 3,5-difluorophenylmethyl as the P1-ligand | | 分子名称: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | 著者 | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | 登録日 | 2017-12-22 | | 公開日 | 2018-02-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Design of Highly Potent, Dual-Acting and Central-Nervous-System-Penetrating HIV-1 Protease Inhibitors with Excellent Potency against Multidrug-Resistant HIV-1 Variants.
ChemMedChem, 13, 2018
|
|
5GVJ
 
 | | Structure of FabK (M276A) mutant from Thermotoga maritima | | 分子名称: | Enoyl-[acyl-carrier-protein] reductase [FMN], SODIUM ION | | 著者 | Kim, E.E, Shin, S.C, Ha, B.H, Moon, J.H. | | 登録日 | 2016-09-06 | | 公開日 | 2017-04-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural and biochemical characterization of FabK from Thermotoga maritima.
Biochem. Biophys. Res. Commun., 482, 2017
|
|
1IVF
 
 | |
6C3K
 
 | |
6TB1
 
 | | Crystal structure of thermostable omega transaminase 6-fold mutant from Pseudomonas jessenii | | 分子名称: | Aspartate aminotransferase family protein, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | 著者 | Capra, N, Rozeboom, H.J, Thunnissen, A.M.W.H, Janssen, D.B. | | 登録日 | 2019-10-31 | | 公開日 | 2020-07-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Robust omega-Transaminases by Computational Stabilization of the Subunit Interface.
Acs Catalysis, 10, 2020
|
|
5GHP
 
 | | Crystal structure of human MTH1(G2K/D120A mutant) in complex with 2-oxo-dATP | | 分子名称: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION | | 著者 | Nakamura, T, Waz, S, Hirata, K, Nakabeppu, Y, Yamagata, Y. | | 登録日 | 2016-06-20 | | 公開日 | 2017-01-04 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.192 Å) | | 主引用文献 | Structural and Kinetic Studies of the Human Nudix Hydrolase MTH1 Reveal the Mechanism for Its Broad Substrate Specificity
J. Biol. Chem., 292, 2017
|
|
3BH4
 
 | | High resolution crystal structure of Bacillus amyloliquefaciens alpha-amylase | | 分子名称: | Alpha-amylase, CALCIUM ION, SODIUM ION | | 著者 | Alikhajeh, J, Khajeh, K, Ranjbar, B, Naderi-Manesh, H, Lin, Y.H, Liu, M.Y, Chen, C.J. | | 登録日 | 2007-11-28 | | 公開日 | 2008-12-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structure of Bacillus amyloliquefaciens alpha-amylase at high resolution: implications for thermal stability.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
5GLL
 
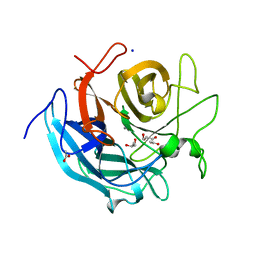 | | Crystal structure of CoXyl43, GH43 beta-xylosidase/alpha-arabinofuranosidase from a compostmicrobial metagenome, calcium-bound form | | 分子名称: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | 著者 | Matsuzawa, T, Kishine, N, Fujimoto, Z, Yaoi, K. | | 登録日 | 2016-07-12 | | 公開日 | 2017-03-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of metagenomic beta-xylosidase/ alpha-l-arabinofuranosidase activated by calcium.
J. Biochem., 162, 2017
|
|
