1LCI
 
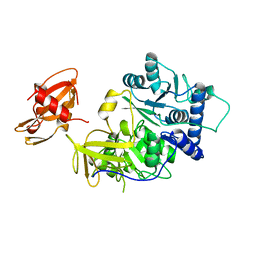 | | FIREFLY LUCIFERASE | | 分子名称: | LUCIFERASE | | 著者 | Conti, E, Franks, N.P, Brick, P. | | 登録日 | 1996-06-01 | | 公開日 | 1997-03-26 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of firefly luciferase throws light on a superfamily of adenylate-forming enzymes.
Structure, 4, 1996
|
|
2GKW
 
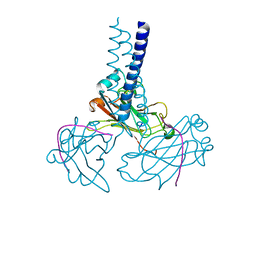 | | Key contacts promote recongnito of BAFF-R by TRAF3 | | 分子名称: | TNF receptor-associated factor 3, Tumor necrosis factor receptor superfamily member 13C | | 著者 | Ely, K.R. | | 登録日 | 2006-04-03 | | 公開日 | 2006-04-11 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Key molecular contacts promote recognition of the BAFF receptor by TNF receptor-associated factor 3: implications for intracellular signaling regulation.
J.Immunol., 173, 2004
|
|
4ETP
 
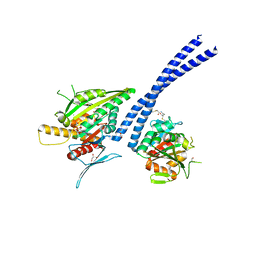 | | C-terminal motor and motor homology domain of Kar3Vik1 fused to a synthetic heterodimeric coiled coil | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, Kinesin-like protein KAR3, ... | | 著者 | Rank, K.C, Chen, C.J, Cope, J, Porche, K, Hoenger, A, Gilbert, S.P, Rayment, I. | | 登録日 | 2012-04-24 | | 公開日 | 2012-06-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Kar3Vik1, a member of the kinesin-14 superfamily, shows a novel kinesin microtubule binding pattern.
J.Cell Biol., 197, 2012
|
|
3CLS
 
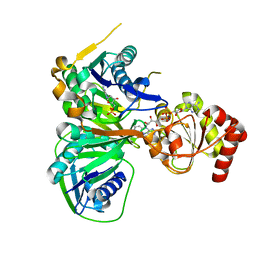 | | Crystal structure of the R236C mutant of ETF from Methylophilus methylotrophus | | 分子名称: | ADENOSINE MONOPHOSPHATE, Electron transfer flavoprotein subunit alpha, Electron transfer flavoprotein subunit beta, ... | | 著者 | Katona, G, Leys, D. | | 登録日 | 2008-03-20 | | 公開日 | 2008-04-08 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Probing the dynamic interface between trimethylamine dehydrogenase (TMADH) and electron transferring flavoprotein (ETF) in the TMADH-2ETF complex: role of the Arg-alpha237 (ETF) and Tyr-442 (TMADH) residue pair.
Biochemistry, 47, 2008
|
|
1GQ4
 
 | |
2F5I
 
 | |
1GQ5
 
 | |
2EZD
 
 | | SOLUTION STRUCTURE OF A COMPLEX OF THE SECOND DNA BINDING DOMAIN OF HUMAN HMG-I(Y) BOUND TO DNA DODECAMER CONTAINING THE PRDII SITE OF THE INTERFERON-BETA PROMOTER, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | DNA (5'-D(*GP*AP*GP*GP*AP*AP*TP*TP*TP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*TP*C)-3'), HIGH MOBILITY GROUP PROTEIN HMG-I/HMG-Y | | 著者 | Clore, G.M, Huth, J.R, Bewley, C, Gronenborn, A.M. | | 登録日 | 1997-06-04 | | 公開日 | 1997-10-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif.
Nat.Struct.Biol., 4, 1997
|
|
1T17
 
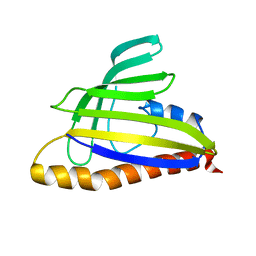 | | Solution Structure of the 18 kDa Protein CC1736 from Caulobacter crescentus: The Northeast Structural Genomics Consortium Target CcR19 | | 分子名称: | conserved hypothetical protein | | 著者 | Shen, Y, Atreya, H.S, Acton, T, Xiao, R, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2004-04-15 | | 公開日 | 2005-01-04 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of the 18 kDa protein CC1736 from Caulobacter crescentus identifies a member of the START domain superfamily and suggests residues mediating substrate specificity.
Proteins, 58, 2005
|
|
2EZE
 
 | | SOLUTION STRUCTURE OF A COMPLEX OF THE SECOND DNA BINDING DOMAIN OF HUMAN HMG-I(Y) BOUND TO DNA DODECAMER CONTAINING THE PRDII SITE OF THE INTERFERON-BETA PROMOTER, NMR, 35 STRUCTURES | | 分子名称: | DNA (5'-D(*GP*AP*GP*GP*AP*AP*TP*TP*TP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*TP*C)-3'), HIGH MOBILITY GROUP PROTEIN HMG-I/HMG-Y | | 著者 | Clore, G.M, Huth, J.R, Bewley, C, Gronenborn, A.M. | | 登録日 | 1997-06-04 | | 公開日 | 1997-10-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif.
Nat.Struct.Biol., 4, 1997
|
|
2EZF
 
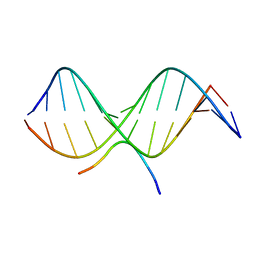 | | SOLUTION STRUCTURE OF A COMPLEX OF THE THIRD DNA BINDING DOMAIN OF HUMAN HMG-I(Y) BOUND TO DNA DODECAMER CONTAINING THE PRDII SITE OF THE INTERFERON-BETA PROMOTER, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | DNA (5'-D(*GP*AP*GP*GP*AP*AP*TP*TP*TP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*TP*C)-3'), HIGH MOBILITY GROUP PROTEIN HMG-I/HMG-Y | | 著者 | Clore, G.M, Huth, J.R, Bewley, C, Gronenborn, A.M. | | 登録日 | 1997-06-04 | | 公開日 | 1997-10-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif.
Nat.Struct.Biol., 4, 1997
|
|
2EZG
 
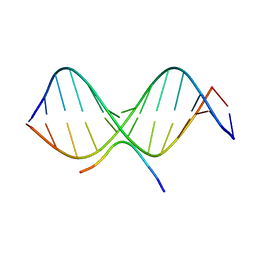 | | SOLUTION STRUCTURE OF A COMPLEX OF THE THIRD DNA BINDING DOMAIN OF HUMAN HMG-I(Y) BOUND TO DNA DODECAMER CONTAINING THE PRDII SITE OF THE INTERFERON-BETA PROMOTER, NMR, 35 STRUCTURES | | 分子名称: | DNA (5'-D(*GP*AP*GP*GP*AP*AP*TP*TP*TP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*TP*C)-3'), HIGH MOBILITY GROUP PROTEIN HMG-I/HMG-Y | | 著者 | Clore, G.M, Huth, J.R, Bewley, C, Gronenborn, A.M. | | 登録日 | 1997-06-04 | | 公開日 | 1997-10-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif.
Nat.Struct.Biol., 4, 1997
|
|
3HZC
 
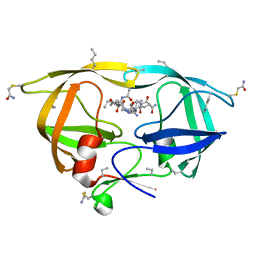 | |
4Z54
 
 | | High Resolution Human Septin3 GTPase domain with alpha-zero helix in complex with GDP | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Neuronal-specific septin-3 | | 著者 | Valadares, N.F, Macedo, J.N, Pereira, H.M, Brandao-Neto, J, Matos, S.O, Leonardo, D.A, Araujo, A.P.U, Garratt, R.C. | | 登録日 | 2015-04-02 | | 公開日 | 2016-04-06 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | A complete compendium of crystal structures for the human SEPT3 subgroup reveals functional plasticity at a specific septin interface
Iucrj, 2020
|
|
4GBY
 
 | | The structure of the MFS (major facilitator superfamily) proton:xylose symporter XylE bound to D-xylose | | 分子名称: | D-xylose-proton symporter, beta-D-xylopyranose, nonyl beta-D-glucopyranoside | | 著者 | Sun, L.F, Zeng, X, Yan, C.Y, Yan, N. | | 登録日 | 2012-07-28 | | 公開日 | 2012-10-17 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.808 Å) | | 主引用文献 | Crystal structure of a bacterial homologue of glucose transporters GLUT1-4.
Nature, 490, 2012
|
|
3HTU
 
 | |
4B3P
 
 | | Structures of HIV-1 RT and RNA-DNA Complex Reveal a Unique RT Conformation and Substrate Interface | | 分子名称: | DNA, P51 RT, REVERSE TRANSCRIPTASE/RIBONUCLEASE H, ... | | 著者 | Lapkouski, M, Tian, L, Miller, J.T, Le Grice, S.F.J, Yang, W. | | 登録日 | 2012-07-25 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (4.839 Å) | | 主引用文献 | Complexes of HIV-1 RT, Nnrti and RNA/DNA Hybrid Reveal a Structure Compatible with RNA Degradation
Nat.Struct.Mol.Biol., 20, 2013
|
|
4B3O
 
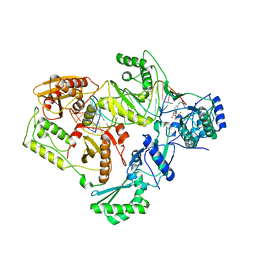 | | Structures of HIV-1 RT and RNA-DNA Complex Reveal a Unique RT Conformation and Substrate Interface | | 分子名称: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, 5'-D(*CP*GP*TP*AP*TP*GP*CP*CP*TP*AP*TP*AP*GP*TP *TP*AP*TP*TP*GP*TP*GP*GP*CP*C)-3', 5'-R(*AP*UP*GP*AP*3DRP*GP*GP*CP*CP*AP*CP*AP*AP*UP*AP *AP*CP*UP*AP*UP*AP*GP*GP*CP*AP*UP*A)-3', ... | | 著者 | Lapkouski, M, Tian, L, Miller, J.T, Le Grice, S.F.J, Yang, W. | | 登録日 | 2012-07-25 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Complexes of HIV-1 RT, Nnrti and RNA/DNA Hybrid Reveal a Structure Compatible with RNA Degradation
Nat.Struct.Mol.Biol., 20, 2013
|
|
4GBZ
 
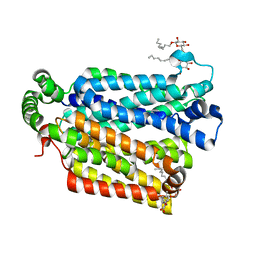 | | The structure of the MFS (major facilitator superfamily) proton:xylose symporter XylE bound to D-glucose | | 分子名称: | D-xylose-proton symporter, beta-D-glucopyranose, nonyl beta-D-glucopyranoside | | 著者 | Sun, L.F, Zeng, X, Yan, C.Y, Yan, N. | | 登録日 | 2012-07-28 | | 公開日 | 2012-10-17 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.894 Å) | | 主引用文献 | Crystal structure of a bacterial homologue of glucose transporters GLUT1-4.
Nature, 490, 2012
|
|
3I2L
 
 | |
4GC0
 
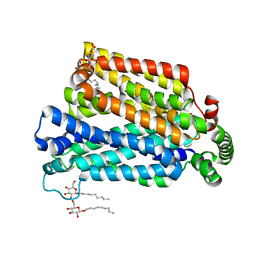 | | The structure of the MFS (major facilitator superfamily) proton:xylose symporter XylE bound to 6-bromo-6-deoxy-D-glucose | | 分子名称: | 6-bromo-6-deoxy-beta-D-glucopyranose, D-xylose-proton symporter, nonyl beta-D-glucopyranoside | | 著者 | Yan, N, Sun, L.F, Zeng, X, Yan, C.Y. | | 登録日 | 2012-07-28 | | 公開日 | 2012-10-17 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of a bacterial homologue of glucose transporters GLUT1-4.
Nature, 490, 2012
|
|
4QXO
 
 | | Crystal structure of hSTING(group2) in complex with DMXAA | | 分子名称: | (5,6-dimethyl-9-oxo-9H-xanthen-4-yl)acetic acid, Stimulator of interferon genes protein | | 著者 | Gao, P, Patel, D.J. | | 登録日 | 2014-07-21 | | 公開日 | 2014-09-10 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Binding-Pocket and Lid-Region Substitutions Render Human STING Sensitive to the Species-Specific Drug DMXAA.
Cell Rep, 8, 2014
|
|
4I5X
 
 | | Crystal Structure Of AKR1B10 Complexed With NADP+ And Flufenamic acid | | 分子名称: | 2-[[3-(TRIFLUOROMETHYL)PHENYL]AMINO] BENZOIC ACID, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Zhang, L, Zheng, X, Chen, S, Zhai, J, Zhang, H, Zhao, Y. | | 登録日 | 2012-11-29 | | 公開日 | 2013-10-23 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Inhibitor selectivity between aldo-keto reductase superfamily members AKR1B10 and AKR1B1: Role of Trp112 (Trp111)
Febs Lett., 587, 2013
|
|
2ILK
 
 | |
4QXQ
 
 | |
