8COO
 
 | | Solution structure of Zipcode binding protein 1 (ZBP1) KH3(DD)KH4 domains in complex with N6-Methyladenosine containing RNA | | 分子名称: | Insulin-like growth factor 2 mRNA-binding protein 1, RNA_(5'-R(*(UP*CP*GP*GP*(6MZ)P*CP*U)-3') | | 著者 | Nicastro, G, Abis, G, Taylor, I.A, Ramos, A. | | 登録日 | 2023-02-28 | | 公開日 | 2024-02-07 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Direct m6A recognition by IMP1 underlays an alternative model of target selection for non-canonical methyl-readers.
Nucleic Acids Res., 51, 2023
|
|
5ZW2
 
 | | FAD complex of PigA | | 分子名称: | 1,2-ETHANEDIOL, 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, ACETATE ION, ... | | 著者 | Lee, C.-C, Ko, T.-P, Wang, A.H.J. | | 登録日 | 2018-05-14 | | 公開日 | 2018-09-05 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.803 Å) | | 主引用文献 | Crystal Structure of PigA: A Prolyl Thioester-Oxidizing Enzyme in Prodigiosin Biosynthesis.
Chembiochem, 20, 2019
|
|
7UOF
 
 | | Dihydroorotase from M. jannaschii | | 分子名称: | Dihydroorotase, ZINC ION | | 著者 | Vitali, J, Nix, J.C, Newman, H.E, Colaneri, M.J. | | 登録日 | 2022-04-12 | | 公開日 | 2022-08-31 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of Methanococcus jannaschii dihydroorotase.
Proteins, 91, 2023
|
|
7UOQ
 
 | | CRYSTAL STRUCTURE OF HIV-1 INTEGRASE COMPLEXED WITH (2S)-2-(TERT-BUTOXY)-2-(5-{2-[(2-CHLORO-6-M ETHYLPHENYL)METHYL]-1,2,3,4-TETRAHYDROISOQUINOLIN-6-YL}-4- (4,4-DIMETHYLPIPERIDIN-1-YL)-2-METHYLPYRIDIN-3-YL)ACETIC ACID | | 分子名称: | (2S)-tert-butoxy[(5M)-5-{2-[(2-chloro-6-methylphenyl)methyl]-1,2,3,4-tetrahydroisoquinolin-6-yl}-4-(4,4-dimethylpiperidin-1-yl)-2-methylpyridin-3-yl]acetic acid, Integrase, SULFATE ION | | 著者 | Lewis, H.A, Muckelbauer, J.K. | | 登録日 | 2022-04-13 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8867 Å) | | 主引用文献 | Discovery and Preclinical Profiling of GSK3839919, a Potent HIV-1 Allosteric Integrase Inhibitor.
Acs Med.Chem.Lett., 13, 2022
|
|
7UP6
 
 | | Crystal structure of C-terminal domain of MSK1 in complex with in covalently bound literature RSK2 inhibitor pyrrolopyrimidine cyanoacrylamide compound 25 (co-crystal) | | 分子名称: | (E)-3-(3-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)phenyl)-2-cyanoacrylamide bound form, OXAMIC ACID, Ribosomal protein S6 kinase alpha-5 | | 著者 | Yano, J.K, Abendroth, J, Hall, A. | | 登録日 | 2022-04-14 | | 公開日 | 2022-08-31 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Discovery and Characterization of a Novel Series of Chloropyrimidines as Covalent Inhibitors of the Kinase MSK1.
Acs Med.Chem.Lett., 13, 2022
|
|
7UP5
 
 | | Crystal structure of C-terminal Domain of MSK1 in complex with covalently bound pyrrolopyrimidine compound 23 (co-crystal) | | 分子名称: | (2M)-6-chloro-2-(5H-pyrrolo[3,2-d]pyrimidin-5-yl)pyridine-3-carbonitrile, IODIDE ION, Ribosomal protein S6 kinase alpha-5 | | 著者 | Yano, J.K, Edwards, T.E, Hall, A. | | 登録日 | 2022-04-14 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Discovery and Characterization of a Novel Series of Chloropyrimidines as Covalent Inhibitors of the Kinase MSK1.
Acs Med.Chem.Lett., 13, 2022
|
|
7UBF
 
 | |
7UBD
 
 | |
7UBI
 
 | |
7UZL
 
 | |
6AFZ
 
 | | Proton pyrophosphatase-E225H mutant | | 分子名称: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Tsai, J.-Y, Li, K.-M, Sun, Y.-J. | | 登録日 | 2018-08-08 | | 公開日 | 2019-04-10 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.483 Å) | | 主引用文献 | Roles of the Hydrophobic Gate and Exit Channel in Vigna radiata Pyrophosphatase Ion Translocation.
J. Mol. Biol., 431, 2019
|
|
7UBC
 
 | |
7UBE
 
 | |
7UBG
 
 | |
5ZT0
 
 | |
7UBH
 
 | |
8DSZ
 
 | | PPARg bound to partial agonist H3B-487 | | 分子名称: | (2R)-2-{5-[(5-{[(1R)-1-(4-tert-butylphenyl)ethyl]carbamoyl}-2,3-dimethyl-1H-indol-1-yl)methyl]-2-chlorophenoxy}propanoic acid, Peroxisome proliferator-activated receptor gamma | | 著者 | Larsen, N.A. | | 登録日 | 2022-07-24 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Biochemical and structural basis for the pharmacological inhibition of nuclear hormone receptor PPAR gamma by inverse agonists.
J.Biol.Chem., 298, 2022
|
|
6AD8
 
 | | Crystal structure of the E148D mutant CLC-ec1 in 50 mM bromide | | 分子名称: | BROMIDE ION, H(+)/Cl(-) exchange transporter ClcA, antibody Fab fragment heavy chain, ... | | 著者 | Lim, H.-H, Park, K. | | 登録日 | 2018-07-31 | | 公開日 | 2019-08-28 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Mutation of external glutamate residue reveals a new intermediate transport state and anion binding site in a CLC Cl-/H+antiporter.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8PXJ
 
 | | Structure of Whitewater Arroyo virus GP1 glycoprotein, solved at wavelength 2.75 A | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, Glycoprotein G1, ... | | 著者 | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Bowden, T.A, Wagner, A. | | 登録日 | 2023-07-23 | | 公開日 | 2023-10-25 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
6ABI
 
 | | The apo-structure of D-lactate dehydrogenase from Fusobacterium nucleatum | | 分子名称: | D-lactate dehydrogenase, GLYCEROL, SULFATE ION | | 著者 | Furukawa, N, Miyanaga, A, Nakajima, M, Taguchi, H. | | 登録日 | 2018-07-21 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Basis of Sequential Allosteric Transitions in Tetrameric d-Lactate Dehydrogenases from Three Gram-Negative Bacteria
Biochemistry, 57, 2018
|
|
8PD8
 
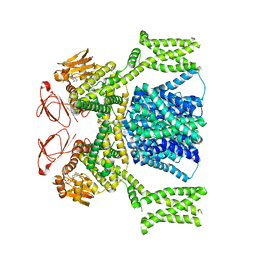 | | cAMP-bound SpSLC9C1 in lipid nanodiscs, dimer | | 分子名称: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Sperm-specific sodium proton exchanger | | 著者 | Kalienkova, V, Peter, M, Rheinberger, J, Paulino, C. | | 登録日 | 2023-06-12 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structures of a sperm-specific solute carrier gated by voltage and cAMP.
Nature, 623, 2023
|
|
8PD3
 
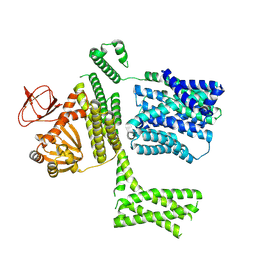 | | Ligand-free SpSLC9C1 in lipid nanodiscs, protomer state 2 | | 分子名称: | Sperm-specific sodium proton exchanger | | 著者 | Kalienkova, V, Peter, M, Rheinberger, J, Paulino, C. | | 登録日 | 2023-06-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structures of a sperm-specific solute carrier gated by voltage and cAMP.
Nature, 623, 2023
|
|
5ZW7
 
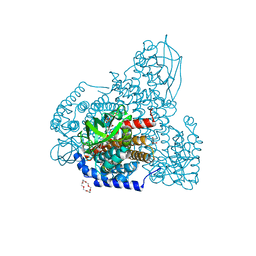 | | FAD-PigA complex at 1.3 A | | 分子名称: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Lee, C.-C, Ko, T.-P, Wang, A.H.J. | | 登録日 | 2018-05-14 | | 公開日 | 2018-09-05 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Crystal Structure of PigA: A Prolyl Thioester-Oxidizing Enzyme in Prodigiosin Biosynthesis.
Chembiochem, 20, 2019
|
|
8DSY
 
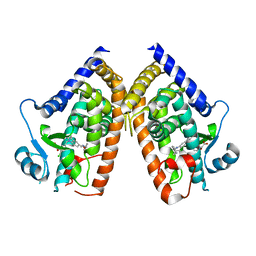 | | PPARg bound to inverse agonist H3B-343 | | 分子名称: | Peroxisome proliferator-activated receptor gamma, {5-[(5-{[(4-tert-butylphenyl)methyl]carbamoyl}-2,3-dimethyl-1H-indol-1-yl)methyl]-2-chlorophenoxy}acetic acid | | 著者 | Larsen, N.A. | | 登録日 | 2022-07-24 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Biochemical and structural basis for the pharmacological inhibition of nuclear hormone receptor PPAR gamma by inverse agonists.
J.Biol.Chem., 298, 2022
|
|
8PD7
 
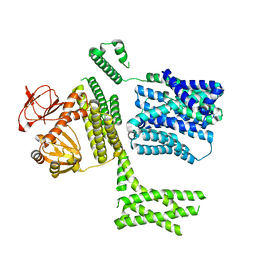 | | Ligand-free SpSLC9C1 in lipid nanodiscs, protomer state 4 | | 分子名称: | Sperm-specific sodium proton exchanger | | 著者 | Kalienkova, V, Peter, M, Rheinberger, J, Paulino, C. | | 登録日 | 2023-06-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structures of a sperm-specific solute carrier gated by voltage and cAMP.
Nature, 623, 2023
|
|
