6C98
 
 | | Crystal structure of FcRn bound to UCB-84 | | 分子名称: | 1-[7-(3-fluorophenyl)-5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-6-yl]ethan-1-one, Beta-2-microglobulin, CYSTEINE, ... | | 著者 | Fox III, D, Lukacs, C.M. | | 登録日 | 2018-01-25 | | 公開日 | 2018-05-30 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Insight into small molecule binding to the neonatal Fc receptor by X-ray crystallography and 100 kHz magic-angle-spinning NMR.
PLoS Biol., 16, 2018
|
|
6C9O
 
 | |
6C99
 
 | | Crystal structure of FcRn bound to UCB-303 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, CITRIC ACID, ... | | 著者 | Fox III, D, Abendroth, J, Porter, J, Deboves, H. | | 登録日 | 2018-01-25 | | 公開日 | 2018-05-30 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Insight into small molecule binding to the neonatal Fc receptor by X-ray crystallography and 100 kHz magic-angle-spinning NMR.
PLoS Biol., 16, 2018
|
|
6C97
 
 | | Crystal structure of FcRn at pH3 | | 分子名称: | Beta-2-microglobulin, GLYCEROL, IgG receptor FcRn large subunit p51 | | 著者 | Fox III, D, Fairman, J.W. | | 登録日 | 2018-01-25 | | 公開日 | 2018-05-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Insight into small molecule binding to the neonatal Fc receptor by X-ray crystallography and 100 kHz magic-angle-spinning NMR.
PLoS Biol., 16, 2018
|
|
6CPZ
 
 | | Selenomethionine mutant (I6Sem) of protein GB1 examined by X-ray diffraction | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Immunoglobulin G-binding protein G, ... | | 著者 | Chen, Q, Rozovsky, S. | | 登録日 | 2018-03-14 | | 公開日 | 2019-07-10 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.12 Å) | | 主引用文献 | 77Se NMR Probes the Protein Environment of Selenomethionine.
J.Phys.Chem.B, 124, 2020
|
|
6CHE
 
 | |
6CNE
 
 | | Selenomethionine variant (V29SeM) of protein GB1 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Immunoglobulin G-binding protein G, PHOSPHATE ION | | 著者 | Chen, Q. | | 登録日 | 2018-03-08 | | 公開日 | 2019-07-10 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | 77Se NMR Probes the Protein Environment of Selenomethionine.
J.Phys.Chem.B, 124, 2020
|
|
7U37
 
 | |
1MA2
 
 | |
7K3G
 
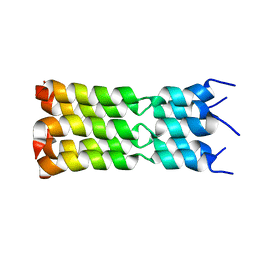 | | SARS-CoV-2 Envelope Protein Transmembrane Domain: Pentameric Structure Determined by Solid-State NMR | | 分子名称: | Envelope small membrane protein | | 著者 | Mandala, V.S, Hong, M, McKay, M.J, Shcherbakov, A.S, Dregni, A.J. | | 登録日 | 2020-09-11 | | 公開日 | 2020-09-30 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | Structure and drug binding of the SARS-CoV-2 envelope protein transmembrane domain in lipid bilayers.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8B29
 
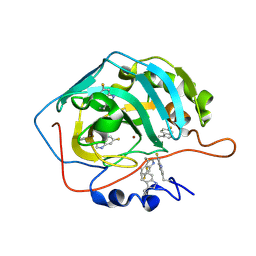 | | Human carbonic anhydrase II containing 6-fluorotryptophanes. | | 分子名称: | Carbonic anhydrase 2, ZINC ION | | 著者 | Pham, L.B.T, Costantino, A, Barbieri, L, Calderone, V, Luchinat, E, Banci, L. | | 登録日 | 2022-09-13 | | 公開日 | 2023-01-18 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Direct Expression of Fluorinated Proteins in Human Cells for 19 F In-Cell NMR Spectroscopy.
J.Am.Chem.Soc., 145, 2023
|
|
7M5T
 
 | |
4AKA
 
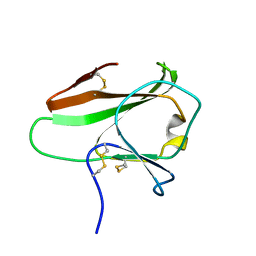 | | IPSE alpha-1, an IgE-binding crystallin | | 分子名称: | IL-4-INDUCING PROTEIN | | 著者 | Meyer, N.H, Mayerhofer, H, Tripsianes, K, Barths, D, Blindow, S, Bade, S, Madl, T, Frey, A, Haas, H, Mueller-Dieckmann, J, Sattler, M, Scharmm, G. | | 登録日 | 2012-02-22 | | 公開日 | 2013-03-13 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A Crystallin Fold in the Interleukin-4-Inducing Principle of Schistosoma Mansoni Eggs (Ipse/Alpha-1) Mediates Ige Binding for Antigen-Independent Basophil Activation
J.Biol.Chem., 290, 2015
|
|
5KK3
 
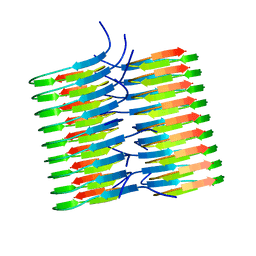 | | Atomic Resolution Structure of Monomorphic AB42 Amyloid Fibrils | | 分子名称: | Beta-amyloid protein 42 | | 著者 | Colvin, M.T, Silvers, R, Zhe Ni, Q, Can, T.V, Sergeyev, I, Rosay, M, Donovan, K.J, Michael, B, Wall, J, Linse, S, Griffin, R.G. | | 登録日 | 2016-06-20 | | 公開日 | 2016-07-13 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | Atomic Resolution Structure of Monomorphic A beta 42 Amyloid Fibrils.
J.Am.Chem.Soc., 138, 2016
|
|
5MPZ
 
 | |
8PSH
 
 | | HIGH RESOLUTION NMR STRUCTURE OF THE STEREOREGULAR (ALL-RP)-PHOSPHOROTHIOATE-DNA/RNA HYBRID D (G*PS*C*PS*G*PS*T*PS*C*PS*A*PS*G*PS*G)R(CCUGACGC), MINIMIZED AVERAGE STRUCTURE | | 分子名称: | DNA (5'-D(*DGP*(SC)P*(GS)P*(PST)P*(SC)P*(AS)P*(GS)P*(GS))-3'), RNA (5'-R(*CP*CP*UP*GP*AP*CP*GP*C)-3') | | 著者 | Bachelin, M, Hessler, G, Kurz, G, Hacia, J.G, Dervan, P.B, Kessler, H. | | 登録日 | 1997-10-13 | | 公開日 | 1998-05-27 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of a Stereoregular Phosphorothioate DNA/RNA Duplex
Nat.Struct.Biol., 5, 1998
|
|
6FGM
 
 | | The NMR solution structure of the peptide AC12 from Hypsiboas raniceps | | 分子名称: | ALA-CYS-PHE-LEU-THR-ARG-LEU-GLY-THR-TYR-VAL-CYS | | 著者 | Popov, C.S.F.C, Simas, B.S, Goodfellow, B.J, Bocca, A.L, Andrade, P.B, Pereira, D, Valentao, P, Pereira, P.J.B, Rodrigues, J.E, Veloso Jr, P.H.H, Rezende, T.M.B. | | 登録日 | 2018-01-11 | | 公開日 | 2019-01-09 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Host-defense peptides AC12, DK16 and RC11 with immunomodulatory activity isolated from Hypsiboas raniceps skin secretion.
Peptides, 113, 2019
|
|
5MQG
 
 | |
5MQE
 
 | |
5MQK
 
 | |
7JQ8
 
 | | Solution NMR structure of human Brd3 ET domain | | 分子名称: | Bromodomain-containing protein 3, Integrase peptide | | 著者 | Aiyer, S, Swapna, G.V.T, Roth, J.M, Montelione, G.T. | | 登録日 | 2020-08-10 | | 公開日 | 2021-06-23 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A common binding motif in the ET domain of BRD3 forms polymorphic structural interfaces with host and viral proteins.
Structure, 29, 2021
|
|
5C5A
 
 | | Crystal Structure of HDM2 in complex with Nutlin-3a | | 分子名称: | 4-({(4S,5R)-4,5-bis(4-chlorophenyl)-2-[4-methoxy-2-(propan-2-yloxy)phenyl]-4,5-dihydro-1H-imidazol-1-yl}carbonyl)piperazin-2-one, CHLORIDE ION, E3 ubiquitin-protein ligase Mdm2, ... | | 著者 | Orts, J, Waelti, M.A, Marsh, M, Vera, L, Gossert, A.D, Guentert, P, Riek, R. | | 登録日 | 2015-06-19 | | 公開日 | 2016-06-29 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.146 Å) | | 主引用文献 | NMR Molecular Replacement, NMR2
To Be Published
|
|
6L7Z
 
 | | Solution NMR structure of the N-terminal immunoglobulin variable domain of BTNL2 | | 分子名称: | Butyrophilin-like protein 2 | | 著者 | Basak, A.J, Lee, W, Samanta, D, De, S. | | 登録日 | 2019-11-03 | | 公開日 | 2020-10-14 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Insights into N-terminal IgV Domain of BTNL2, a T Cell Inhibitory Molecule, Suggests a Non-canonical Binding Interface for Its Putative Receptors.
J.Mol.Biol., 432, 2020
|
|
5LWJ
 
 | | Solution NMR structure of the GTP binding Class II RNA aptamer-ligand-complex containing a protonated adenine nucleotide with a highly shifted pKa. | | 分子名称: | GTP Class II RNA (34-MER), GUANOSINE-5'-TRIPHOSPHATE | | 著者 | Wolter, A.C, Weickhmann, A.K, Nasiri, A.H, Hantke, K, Ohlenschlaeger, O, Wunderlich, C.H, Kreutz, C, Duchardt-Ferner, E, Woehnert, J. | | 登録日 | 2016-09-17 | | 公開日 | 2016-12-14 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A Stably Protonated Adenine Nucleotide with a Highly Shifted pKa Value Stabilizes the Tertiary Structure of a GTP-Binding RNA Aptamer.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
3MOM
 
 | | Structure of holo HasAp H32A mutant complexed with imidazole from Pseudomonas aeruginosa to 2.25A Resolution | | 分子名称: | Heme acquisition protein HasAp, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Lovell, S, Battaile, K.P, Jepkorir, G, Rodriguez, J.C, Rui, H, Im, W, Alontaga, A.Y, Yukl, E, Moenne-Loccoz, P, Rivera, M. | | 登録日 | 2010-04-22 | | 公開日 | 2010-07-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structural, NMR Spectroscopic, and Computational Investigation of Hemin Loading in the Hemophore HasAp from Pseudomonas aeruginosa.
J.Am.Chem.Soc., 132, 2010
|
|
