7QP2
 
 | | 1-deazaguanosine modified-RNA Sarcin Ricin Loop | | 分子名称: | GLYCEROL, RNA (27-MER) | | 著者 | Ennifar, E, Micura, R, Bereiter, R, Renard, E, Kreutz, C. | | 登録日 | 2021-12-30 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | 1-Deazaguanosine-Modified RNA: The Missing Piece for Functional RNA Atomic Mutagenesis.
J.Am.Chem.Soc., 144, 2022
|
|
1VYR
 
 | | Structure of pentaerythritol tetranitrate reductase complexed with picric acid | | 分子名称: | FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE, PICRIC ACID | | 著者 | Barna, T, Moody, P.C.E. | | 登録日 | 2004-05-05 | | 公開日 | 2004-06-10 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Atomic Resolution Structures and Solution Behavior of Enzyme-Substrate Complexes of Enterobacter Cloacae Pb2 Pentaerythritol Tetranitrate Reductase: Multiple Conformational States and Implications for the Mechanism of Nitroaromatic Explosive Degradation
J.Biol.Chem., 279, 2004
|
|
6B00
 
 | |
8DDG
 
 | |
6Y14
 
 | |
6M9J
 
 | | Racemic-GSTSTA from degenerate octameric repeats in InaZ, residues 707-712 | | 分子名称: | Ice nucleation protein | | 著者 | Zee, C, Glynn, C, Gallagher-Jones, M, Miao, J, Santiago, C.G, Cascio, D, Gonen, T, Sawaya, M.R, Rodriguez, J.A. | | 登録日 | 2018-08-23 | | 公開日 | 2019-03-27 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (0.9 Å) | | 主引用文献 | Homochiral and racemic MicroED structures of a peptide repeat from the ice-nucleation protein InaZ.
IUCrJ, 6, 2019
|
|
1OEW
 
 | | ATOMIC RESOLUTION STRUCTURE OF NATIVE ENDOTHIAPEPSIN | | 分子名称: | ENDOTHIAPEPSIN, GLYCEROL, SERINE, ... | | 著者 | Coates, L, Erskine, P.T, Mall, S, Gill, R.S, Wood, S.P, Myles, D.A.A, Cooper, J.B. | | 登録日 | 2003-03-31 | | 公開日 | 2003-04-02 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Atomic Resolution Analysis of the Catalytic Site of an Aspartic Proteinase and an Unexpected Mode of Binding by Short Peptides
Protein Sci., 12, 2003
|
|
6ZYB
 
 | |
7WDJ
 
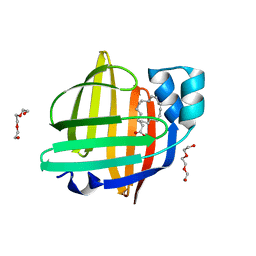 | | The 0.90 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with linoleic acid | | 分子名称: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | 著者 | Sugiyama, S, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | 登録日 | 2021-12-21 | | 公開日 | 2022-12-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | The 0.90 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with linoleic acid
To Be Published
|
|
7TWT
 
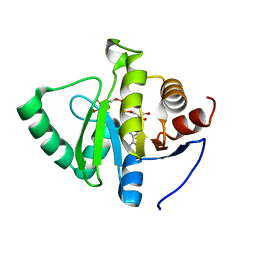 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ADP-ribose at pH 4 (P43 crystal form) | | 分子名称: | Non-structural protein 3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | 著者 | Correy, G.J, Fraser, J.S. | | 登録日 | 2022-02-07 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ADP-ribose at pH 4 (P43 crystal form)
To Be Published
|
|
2WUR
 
 | |
5Y2S
 
 | |
6BZM
 
 | | GFGNFGTS from low-complexity/FG repeat domain of Nup98, residues 116-123 | | 分子名称: | Nuclear pore complex protein Nup98-Nup96 | | 著者 | Hughes, M.P, Rodriguez, J.A, Sawaya, M.R, Cascio, D, Chong, L, Gonen, T, Eisenberg, D.S. | | 登録日 | 2017-12-24 | | 公開日 | 2018-04-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (0.9 Å) | | 主引用文献 | Atomic structures of low-complexity protein segments reveal kinked beta sheets that assemble networks.
Science, 359, 2018
|
|
1N9B
 
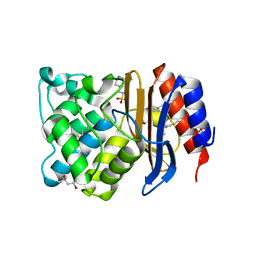 | | Ultrahigh resolution structure of a class A beta-lactamase: On the mechanism and specificity of the extended-spectrum SHV-2 enzyme | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BETA-LACTAMASE SHV-2, ... | | 著者 | Nukaga, M, Mayama, K, Hujer, A.M, Bonomo, R.A, Knox, J.R. | | 登録日 | 2002-11-22 | | 公開日 | 2003-04-08 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Ultrahigh resolution structure of a class A beta-lactamase: On the mechanism and specificity of the extended-spectrum SHV-2 enzyme
J.Mol.Biol., 328, 2003
|
|
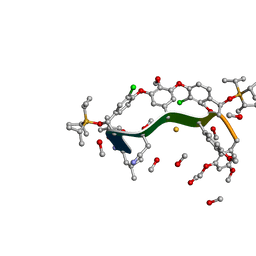
8GJV
 
 | |
6XVM
 
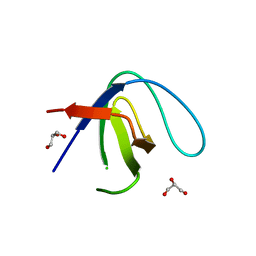 | |
3G21
 
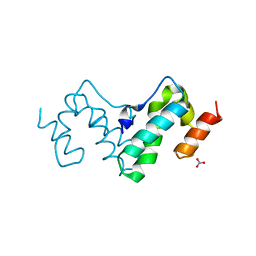 | |
4EA9
 
 | | X-ray structure of GDP-perosamine N-acetyltransferase in complex with transition state analog at 0.9 Angstrom resolution | | 分子名称: | CHLORIDE ION, GDP-N-acetylperosamine-coenzyme A, Perosamine N-acetyltransferase | | 著者 | Thoden, J.B, Reinhardt, L.A, Cook, P.D, Menden, P, Cleland, W.W, Holden, H.M. | | 登録日 | 2012-03-22 | | 公開日 | 2012-04-04 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Catalytic Mechanism of Perosamine N-Acetyltransferase Revealed by High-Resolution X-ray Crystallographic Studies and Kinetic Analyses.
Biochemistry, 51, 2012
|
|
1G66
 
 | | ACETYLXYLAN ESTERASE AT 0.90 ANGSTROM RESOLUTION | | 分子名称: | ACETYL XYLAN ESTERASE II, GLYCEROL, SULFATE ION | | 著者 | Ghosh, D, Sawicki, M, Lala, P, Erman, M, Pangborn, W, Eyzaguirre, J, Gutierrez, R, Jornvall, H, Thiel, D.J. | | 登録日 | 2000-11-03 | | 公開日 | 2001-01-17 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Multiple conformations of catalytic serine and histidine in acetylxylan esterase at 0.90 A.
J.Biol.Chem., 276, 2001
|
|
8ANM
 
 | |
3KS3
 
 | |
5B28
 
 | | The 0.90A structure of human FABP3 F16V mutant complexed with palmitic acid | | 分子名称: | Fatty acid-binding protein, heart, PALMITIC ACID, ... | | 著者 | Matsuoka, D, Sugiyama, S, Kakinouchi, K, Niiyama, M, Murata, M, Matsuoka, S. | | 登録日 | 2016-01-12 | | 公開日 | 2017-01-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | The 0.90A structure of human FABP3 F16V mutant complexed with palmitic acid.
To Be Published
|
|
3ZR8
 
 | | Crystal structure of RxLR effector Avr3a11 from Phytophthora capsici | | 分子名称: | AVR3A11, CHLORIDE ION, TRIETHYLENE GLYCOL | | 著者 | Boutemy, L.S, King, S.R.F, Win, J, Hughes, R.K, Clarke, T.A, Blumenschein, T.M.A, Kamoun, S, Banfield, M.J. | | 登録日 | 2011-06-15 | | 公開日 | 2011-08-03 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Structures of Phytophthora Rxlr Effector Proteins: A Conserved But Adaptable Fold Underpins Functional Diversity.
J.Biol.Chem., 286, 2011
|
|
2BW4
 
 | | Atomic Resolution Structure of Resting State of the Achromobacter cycloclastes Cu Nitrite Reductase | | 分子名称: | ACETATE ION, COPPER (II) ION, COPPER-CONTAINING NITRITE REDUCTASE, ... | | 著者 | Antonyuk, S.V, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | 登録日 | 2005-07-12 | | 公開日 | 2005-08-17 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Atomic Resolution Structures of Resting-State, Substrate- and Product-Complexed Cu-Nitrite Reductase Provide Insight Into Catalytic Mechanism
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1O56
 
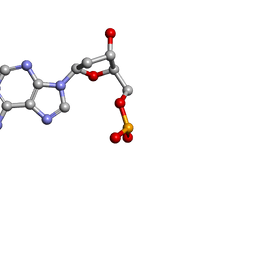 | | MOLECULAR STRUCTURE OF TWO CRYSTAL FORMS OF CYCLIC TRIADENYLIC ACID AT 1 ANGSTROM RESOLUTION | | 分子名称: | DNA (5'-CD(*AP*AP*AP*)-3') | | 著者 | Gao, Y.G, Robinson, H, Guan, Y, Liaw, Y.C, van Boom, J.H, van der Marel, G.A, Wang, A.H. | | 登録日 | 2003-08-20 | | 公開日 | 2003-08-26 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Molecular structure of two crystal forms of cyclic triadenylic acid at 1A resolution.
J.Biomol.Struct.Dyn., 16, 1998
|
|
