4ZV2
 
 | |
6XQS
 
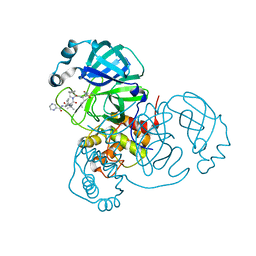 | | Room-temperature X-ray Crystal structure of SARS-CoV-2 main protease in complex with Telaprevir | | 分子名称: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, 3C-like proteinase | | 著者 | Kneller, D.W, Kovalevsky, A, Coates, L. | | 登録日 | 2020-07-10 | | 公開日 | 2020-07-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Malleability of the SARS-CoV-2 3CL M pro Active-Site Cavity Facilitates Binding of Clinical Antivirals.
Structure, 28, 2020
|
|
6L6M
 
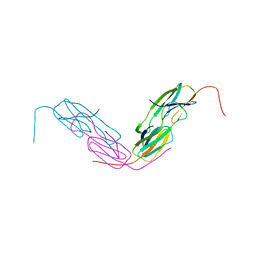 | | HSP18.5 from E. histolytica | | 分子名称: | Heat shock protein hsp20 family putative | | 著者 | Kurre, D, Suguna, K. | | 登録日 | 2019-10-29 | | 公開日 | 2020-11-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.28000379 Å) | | 主引用文献 | Network of Entamoeba histolytica HSP18.5 dimers formed by two overlapping [IV]-X-[IV] motifs.
Proteins, 2021
|
|
6L7X
 
 | |
5ADF
 
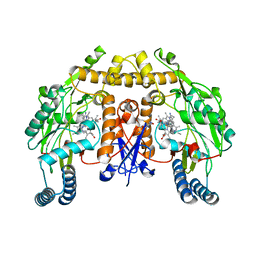 | | Structure of human nNOS R354A G357D mutant heme domain in complex with 7-(((3-((Dimethylamino)methyl)phenyl)amino)methyl)quinolin-2- amine | | 分子名称: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[[[3-[(dimethylamino)methyl]phenyl]amino]methyl]quinolin-2-amine, NITRIC OXIDE SYNTHASE, ... | | 著者 | Li, H, Poulos, T.L. | | 登録日 | 2015-08-20 | | 公開日 | 2015-10-28 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.966 Å) | | 主引用文献 | Phenyl Ether- and Aniline-Containing 2-Aminoquinolines as Potent and Selective Inhibitors of Neuronal Nitric Oxide Synthase.
J.Med.Chem., 58, 2015
|
|
6L36
 
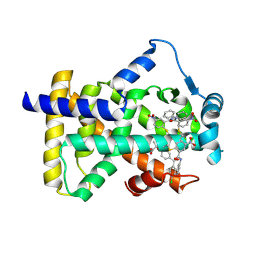 | | X-ray structure of human PPARalpha ligand binding domain-GW9662-fenofibric acid co-crystals obtained by delipidation and co-crystallization | | 分子名称: | 2-[4-(4-chlorobenzene-1-carbonyl)phenoxy]-2-methylpropanoic acid, 2-chloro-5-nitro-N-phenylbenzamide, Peroxisome proliferator-activated receptor alpha | | 著者 | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | 登録日 | 2019-10-09 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.301 Å) | | 主引用文献 | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
4ZVW
 
 | |
6XH3
 
 | |
6L46
 
 | | High-resolution neutron and X-ray joint refined structure of copper-containing nitrite reductase from Geobacillus thermodenitrificans | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, COPPER (II) ION, ... | | 著者 | Fukuda, Y, Hirano, Y, Kusaka, K, Inoue, T, Tamada, T. | | 登録日 | 2019-10-16 | | 公開日 | 2020-02-12 | | 最終更新日 | 2024-04-03 | | 実験手法 | NEUTRON DIFFRACTION (1.3 Å), X-RAY DIFFRACTION | | 主引用文献 | High-resolution neutron crystallography visualizes an OH-bound resting state of a copper-containing nitrite reductase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4ZXR
 
 | | Structure of Thaumatin wrapped in graphene within vacuum | | 分子名称: | GLYCEROL, L(+)-TARTARIC ACID, Thaumatin-1 | | 著者 | Aller, P, Warren, A.J, Trincao, J, Evans, G. | | 登録日 | 2015-05-20 | | 公開日 | 2015-10-14 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | In vacuo X-ray data collection from graphene-wrapped protein crystals.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7DEQ
 
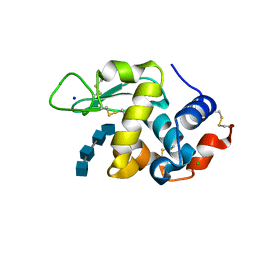 | | Lysozyme-sugar complex in D2O | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | 著者 | Tanaka, I, Chatake, T. | | 登録日 | 2020-11-04 | | 公開日 | 2021-03-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.03 Å) | | 主引用文献 | Recent structural insights into the mechanism of lysozyme hydrolysis.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
4ZT2
 
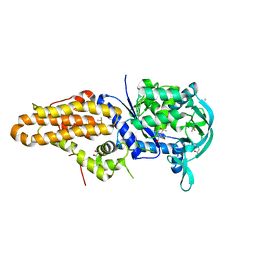 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor N-(3,5-dichlorobenzyl)-N'-(1H-imidazo[4,5-b]pyridin-2-yl)propane-1,3-diamine (Chem 1575) | | 分子名称: | DIMETHYL SULFOXIDE, GLYCEROL, METHIONINE, ... | | 著者 | Koh, C.-Y, Hol, W.G.J. | | 登録日 | 2015-05-14 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | 5-Fluoroimidazo[4,5-b]pyridine Is a Privileged Fragment That Conveys Bioavailability to Potent Trypanosomal Methionyl-tRNA Synthetase Inhibitors.
Acs Infect Dis., 2, 2016
|
|
6XNS
 
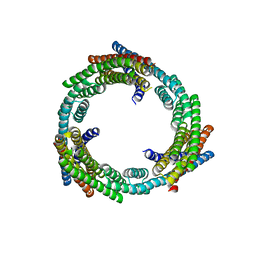 | | C3_crown-05 | | 分子名称: | C3_crown-05 | | 著者 | Bick, M.J, Hsia, Y, Sankaran, B, Baker, D. | | 登録日 | 2020-07-04 | | 公開日 | 2020-12-23 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.19 Å) | | 主引用文献 | Design of multi-scale protein complexes by hierarchical building block fusion.
Nat Commun, 12, 2021
|
|
6LBB
 
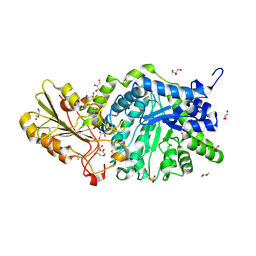 | | Crystal structure of barley exohydrolaseI W434A mutant in complex with 4I,4III,4V-S-trithiocellohexaose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Beta-D-glucan exohydrolase isoenzyme ExoI, ... | | 著者 | Luang, S, Streltsov, V.A, Hrmova, M. | | 登録日 | 2019-11-14 | | 公開日 | 2020-09-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6XP4
 
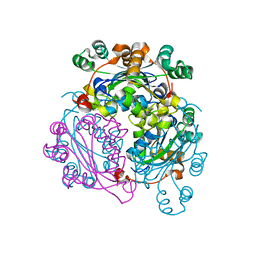 | |
4ZUK
 
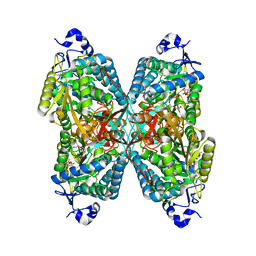 | | Structure ALDH7A1 complexed with NAD+ | | 分子名称: | Alpha-aminoadipic semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TETRAETHYLENE GLYCOL | | 著者 | Luo, M, Tanner, J.J. | | 登録日 | 2015-05-16 | | 公開日 | 2015-08-26 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.001 Å) | | 主引用文献 | Structural Basis of Substrate Recognition by Aldehyde Dehydrogenase 7A1.
Biochemistry, 54, 2015
|
|
6XCA
 
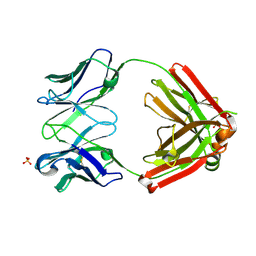 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment, C105 | | 分子名称: | C105 Heavy Chain, C105 Light Chain, SULFATE ION | | 著者 | Sharaf, N.G, Barnes, C.O, Bjorkman, P.J. | | 登録日 | 2020-06-08 | | 公開日 | 2020-07-01 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies.
Cell, 182, 2020
|
|
6LBV
 
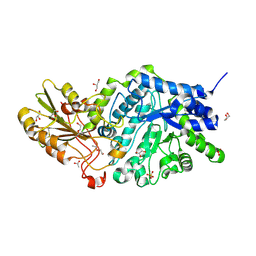 | | Crystal structure of barley exohydrolaseI W434F mutant in complex with methyl 6-thio-beta-gentiobioside | | 分子名称: | ACETATE ION, Beta-D-glucan exohydrolase isoenzyme ExoI, GLYCEROL, ... | | 著者 | Luang, S, Streltsov, V.A, Hrmova, M. | | 登録日 | 2019-11-15 | | 公開日 | 2020-09-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6LCF
 
 | |
6XCC
 
 | |
6L5V
 
 | | Carbonmonoxy human hemoglobin C in the R quaternary structure at 95 K: Light (2 min) | | 分子名称: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | 著者 | Shibayama, N, Park, S.Y, Ohki, M, Sato-Tomita, A. | | 登録日 | 2019-10-24 | | 公開日 | 2020-02-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Direct observation of ligand migration within human hemoglobin at work.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6XPS
 
 | |
7DER
 
 | | Lysozyme alone in H2O | | 分子名称: | CHLORIDE ION, Lysozyme C, SODIUM ION | | 著者 | Tanaka, I, Chatake, T. | | 登録日 | 2020-11-04 | | 公開日 | 2021-03-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.03 Å) | | 主引用文献 | Recent structural insights into the mechanism of lysozyme hydrolysis.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7D98
 
 | |
6XQU
 
 | |
