7VKW
 
 | | The apo structure of beta-1,2-glucosyltransferase from Ignavibacterium album | | 分子名称: | beta-1,2-glucosyltransferase | | 著者 | Kobayashi, K, Shimizu, H, Tanaka, N, Kuramochi, K, Nakai, H, Nakajima, M, Taguchi, H. | | 登録日 | 2021-10-01 | | 公開日 | 2022-03-09 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Characterization and structural analyses of a novel glycosyltransferase acting on the beta-1,2-glucosidic linkages.
J.Biol.Chem., 298, 2022
|
|
7VL6
 
 | | The complex structure of beta-1,2-glucosyltransferase from Ignavibacterium album with arbutin | | 分子名称: | (2R,3S,4S,5R,6S)-2-(hydroxymethyl)-6-(4-oxidanylphenoxy)oxane-3,4,5-triol, CALCIUM ION, beta-1,2-glucosyltransferase | | 著者 | Kobayashi, K, Shimizu, H, Tanaka, N, Kuramochi, K, Nakai, H, Nakajima, M, Taguchi, H. | | 登録日 | 2021-10-01 | | 公開日 | 2022-03-09 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Characterization and structural analyses of a novel glycosyltransferase acting on the beta-1,2-glucosidic linkages.
J.Biol.Chem., 298, 2022
|
|
7VL0
 
 | | The complex structure of beta-1,2-glucosyltransferase from Ignavibacterium album with p-nitrophenyl-alpha-D-glucopyranoside | | 分子名称: | 4-nitrophenyl alpha-D-glucopyranoside, Beta-galactosidase, CALCIUM ION | | 著者 | Kobayashi, K, Shimizu, H, Tanaka, N, Kuramochi, K, Nakai, H, Nakajima, M, Taguchi, H. | | 登録日 | 2021-10-01 | | 公開日 | 2022-03-09 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Characterization and structural analyses of a novel glycosyltransferase acting on the beta-1,2-glucosidic linkages.
J.Biol.Chem., 298, 2022
|
|
7VKZ
 
 | | The complex structure of beta-1,2-glucosyltransferase from Ignavibacterium album with 1-Deoxynojirimycin | | 分子名称: | 1-DEOXYNOJIRIMYCIN, CALCIUM ION, beta-1,2-glucosyltransferase | | 著者 | Kobayashi, K, Shimizu, H, Tanaka, N, Kuramochi, K, Nakai, H, Nakajima, M, Taguchi, H. | | 登録日 | 2021-10-01 | | 公開日 | 2022-03-09 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Characterization and structural analyses of a novel glycosyltransferase acting on the beta-1,2-glucosidic linkages.
J.Biol.Chem., 298, 2022
|
|
7VL3
 
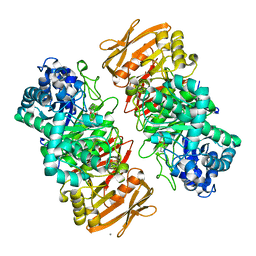 | | The complex structure of beta-1,2-glucosyltransferase from Ignavibacterium album with phenyl alpha-D-glucoside | | 分子名称: | (2R,3S,4S,5R,6R)-2-(hydroxymethyl)-6-phenoxy-oxane-3,4,5-triol, CALCIUM ION, beta-1,2-glucosyltransferase | | 著者 | Kobayashi, K, Shimizu, H, Tanaka, N, Kuramochi, K, Nakai, H, Nakajima, M, Taguchi, H. | | 登録日 | 2021-10-01 | | 公開日 | 2022-03-09 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Characterization and structural analyses of a novel glycosyltransferase acting on the beta-1,2-glucosidic linkages.
J.Biol.Chem., 298, 2022
|
|
7VL4
 
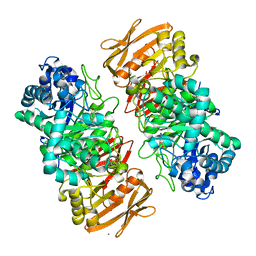 | | The complex structure of beta-1,2-glucosyltransferase from Ignavibacterium album with methyl beta-D-glucoside | | 分子名称: | CALCIUM ION, beta-1,2-glucosyltransferase, methyl beta-D-glucopyranoside | | 著者 | Kobayashi, K, Shimizu, H, Tanaka, N, Kuramochi, K, Nakai, H, Nakajima, M, Taguchi, H. | | 登録日 | 2021-10-01 | | 公開日 | 2022-03-09 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Characterization and structural analyses of a novel glycosyltransferase acting on the beta-1,2-glucosidic linkages.
J.Biol.Chem., 298, 2022
|
|
7VL1
 
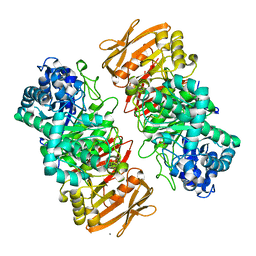 | | The complex structure of beta-1,2-glucosyltransferase from Ignavibacterium album with methyl alpha-D-glucoside | | 分子名称: | CALCIUM ION, beta-1,2-glucosyltransferase, methyl alpha-D-glucopyranoside | | 著者 | Kobayashi, K, Shimizu, H, Tanaka, N, Kuramochi, K, Nakai, H, Nakajima, M, Taguchi, H. | | 登録日 | 2021-10-01 | | 公開日 | 2022-03-09 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Characterization and structural analyses of a novel glycosyltransferase acting on the beta-1,2-glucosidic linkages.
J.Biol.Chem., 298, 2022
|
|
3J8V
 
 | |
7WZO
 
 | |
3F52
 
 | | Crystal structure of the clp gene regulator ClgR from C. glutamicum | | 分子名称: | GLYCEROL, clp gene regulator (ClgR) | | 著者 | Russo, S, Schweitzer, J.E, Polen, T, Bott, M, Pohl, E. | | 登録日 | 2008-11-03 | | 公開日 | 2008-11-18 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal structure of the caseinolytic protease gene regulator, a transcriptional activator in actinomycetes
J.Biol.Chem., 284, 2009
|
|
2C25
 
 | | 1.8A Crystal Structure of Psathyrella velutina lectin in complex with N-acetylneuraminic acid | | 分子名称: | CALCIUM ION, N-acetyl-alpha-neuraminic acid, PSATHYRELLA VELUTINA LECTIN PVL, ... | | 著者 | Cioci, G, Mitchell, E.P, Chazalet, V, Gautier, C, Oscarson, S, Debray, H, Perez, S, Imberty, A. | | 登録日 | 2005-09-26 | | 公開日 | 2006-01-23 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Beta-Propeller Crystal Structure of Psathyrella Velutina Lectin: An Integrin-Like Fungal Protein Interacting with Monosaccharides and Calcium.
J.Mol.Biol., 357, 2006
|
|
7WN2
 
 | |
7WNB
 
 | |
7BB5
 
 | |
3J8Z
 
 | |
3J8W
 
 | |
3F51
 
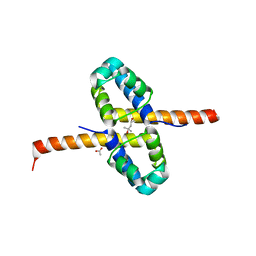 | | Crystal Structure of the clp gene regulator ClgR from Corynebacterium glutamicum | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Clp gene regulator (ClgR) | | 著者 | Russo, S, Schweitzer, J.E, Polen, T, Bott, M, Pohl, E. | | 登録日 | 2008-11-03 | | 公開日 | 2008-11-18 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Crystal structure of the caseinolytic protease gene regulator, a transcriptional activator in actinomycetes
J.Biol.Chem., 284, 2009
|
|
7X87
 
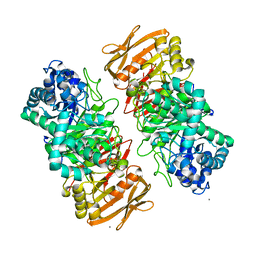 | | The complex structure of beta-1,2-glucosyltransferase from Ignavibacterium album with sophotetraose observed as sophorose | | 分子名称: | Beta-galactosidase, CALCIUM ION, beta-D-glucopyranose-(1-2)-beta-D-glucopyranose | | 著者 | Kobayashi, K, Shimizu, H, Tanaka, N, Kuramochi, K, Nakai, H, Nakajima, M, Taguchi, H. | | 登録日 | 2022-03-11 | | 公開日 | 2022-04-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Characterization and structural analyses of a novel glycosyltransferase acting on the beta-1,2-glucosidic linkages.
J Biol Chem, 298, 2022
|
|
4BXU
 
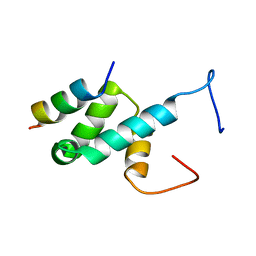 | | Structure of Pex14 in complex with Pex5 LVxEF motif | | 分子名称: | PEROXISOMAL MEMBRANE PROTEIN PEX14, PEROXISOMAL TARGETING SIGNAL 1 RECEPTOR | | 著者 | Kooshapur, H, Meyer, H.N, Madl, T, Sattler, M. | | 登録日 | 2013-07-15 | | 公開日 | 2013-11-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A Novel Pex14 Interacting Site of Human Pex5 is Critical for Matrix Protein Import Into Peroxisomes.
J.Biol.Chem., 289, 2014
|
|
2LT1
 
 | |
7Q21
 
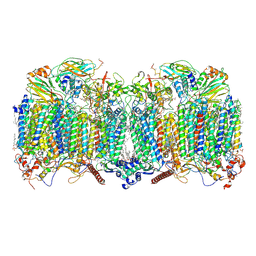 | | III2-IV2 respiratory supercomplex from Corynebacterium glutamicum | | 分子名称: | (1R)-2-(dodecanoyloxy)-1-[(phosphonooxy)methyl]ethyl tetradecanoate, (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2S)-1-(hexadecanoyloxy)propan-2-yl (10S)-10-methyloctadecanoate, ... | | 著者 | Kovalova, T, Moe, A, Krol, S, Yanofsky, D.J, Bott, M, Sjostrand, D, Rubinstein, J.L, Hogbom, M, Brzezinski, P. | | 登録日 | 2021-10-22 | | 公開日 | 2022-02-02 | | 最終更新日 | 2022-03-16 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | The respiratory supercomplex from C. glutamicum.
Structure, 30, 2022
|
|
2MD8
 
 | |
2BIT
 
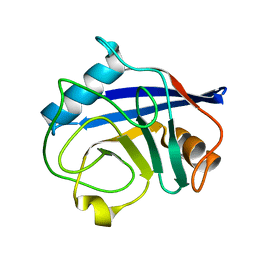 | | Crystal structure of human cyclophilin D at 1.7 A resolution | | 分子名称: | PEPTIDYL-PROLYL CIS-TRANS ISOMERASE | | 著者 | Hennig, M, Thoma, R, Stihle, M, Schlatter, D. | | 登録日 | 2005-01-26 | | 公開日 | 2005-01-26 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Crystal Engineering Yields Crystals of Cyclophilin D Diffracting to 1.7 A Resolution
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2MD7
 
 | |
2CHU
 
 | | CeuE in complex with mecam | | 分子名称: | ENTEROCHELIN UPTAKE PERIPLASMIC BINDING PROTEIN, FE (III) ION, N,N',N''-[BENZENE-1,3,5-TRIYLTRIS(METHYLENE)]TRIS(2,3-DIHYDROXYBENZAMIDE), ... | | 著者 | Muller, A, Wilkinson, A.J, Wilson, K.S, Duhme-Klair, A.K. | | 登録日 | 2006-03-16 | | 公開日 | 2006-08-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | An [{Fe(Mecam)}(2)](6-) Bridge in the Crystal Structure of a Ferric Enterobactin Binding Protein.
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
