1CQT
 
 | | CRYSTAL STRUCTURE OF A TERNARY COMPLEX CONTAINING AN OCA-B PEPTIDE, THE OCT-1 POU DOMAIN, AND AN OCTAMER ELEMENT | | 分子名称: | DNA (5'-D(*AP*CP*CP*TP*TP*AP*TP*TP*TP*GP*CP*AP*TP*AP*C)-3'), DNA (5'-D(*TP*GP*TP*AP*TP*GP*CP*AP*AP*AP*TP*AP*AP*GP*G)-3'), POU DOMAIN, ... | | 著者 | Chasman, D.I, Cepek, K, Sharp, P.A, Pabo, C.O. | | 登録日 | 1999-08-11 | | 公開日 | 1999-11-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Crystal structure of an OCA-B peptide bound to an Oct-1 POU domain/octamer DNA complex: specific recognition of a protein-DNA interface.
Genes Dev., 13, 1999
|
|
4E2A
 
 | |
1F89
 
 | | Crystal structure of Saccharomyces cerevisiae Nit3, a member of branch 10 of the nitrilase superfamily | | 分子名称: | 32.5 KDA PROTEIN YLR351C | | 著者 | Kumaran, D, Eswaramoorthy, S, Studier, F.W, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2000-06-29 | | 公開日 | 2001-10-04 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of a putative CN hydrolase from yeast
Proteins, 52, 2003
|
|
4NTN
 
 | | E.coli QueD, SeMet protein, 2A resolution | | 分子名称: | 6-carboxy-5,6,7,8-tetrahydropterin synthase, FORMIC ACID, ZINC ION | | 著者 | Bandarian, V, Roberts, S.A, Miles, Z.D. | | 登録日 | 2013-12-02 | | 公開日 | 2014-07-16 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Biochemical and Structural Studies of 6-Carboxy-5,6,7,8-tetrahydropterin Synthase Reveal the Molecular Basis of Catalytic Promiscuity within the Tunnel-fold Superfamily.
J.Biol.Chem., 289, 2014
|
|
4NTK
 
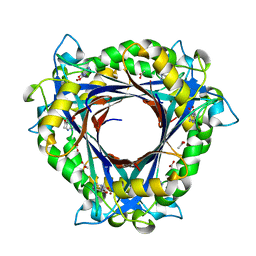 | | QueD from E. coli | | 分子名称: | 2-amino-6-[(1Z)-1,2-dihydroxyprop-1-en-1-yl]-7,8-dihydropteridin-4(3H)-one, 6-carboxy-5,6,7,8-tetrahydropterin synthase, ACETATE ION, ... | | 著者 | Bandarian, V, Roberts, S.A, Miles, Z.D. | | 登録日 | 2013-12-02 | | 公開日 | 2014-07-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Biochemical and Structural Studies of 6-Carboxy-5,6,7,8-tetrahydropterin Synthase Reveal the Molecular Basis of Catalytic Promiscuity within the Tunnel-fold Superfamily.
J.Biol.Chem., 289, 2014
|
|
4NTM
 
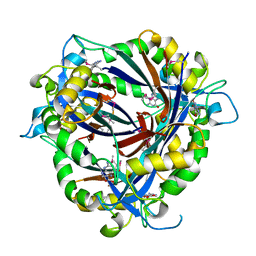 | | QueD soaked with sepiapterin (selenomethionine substituted protein) | | 分子名称: | (6R)-2-amino-4-oxo-3,4,5,6,7,8-hexahydropteridine-6-carboxylic acid, 6-carboxy-5,6,7,8-tetrahydropterin synthase, ZINC ION | | 著者 | Bandarian, V, Miles, Z.D, Roberts, S.A. | | 登録日 | 2013-12-02 | | 公開日 | 2014-07-16 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Biochemical and Structural Studies of 6-Carboxy-5,6,7,8-tetrahydropterin Synthase Reveal the Molecular Basis of Catalytic Promiscuity within the Tunnel-fold Superfamily.
J.Biol.Chem., 289, 2014
|
|
3U4K
 
 | |
6KLW
 
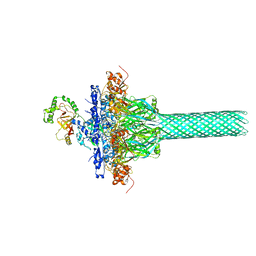 | | Complex structure of Iota toxin enzymatic component (Ia) and binding component (Ib) pore with long stem | | 分子名称: | CALCIUM ION, Iota toxin component Ia, Iota toxin component Ib | | 著者 | Yoshida, T, Yamada, T, Kawamoto, A, Mitsuoka, K, Iwasaki, K, Tsuge, H. | | 登録日 | 2019-07-30 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Cryo-EM structures reveal translocational unfolding in the clostridial binary iota toxin complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5CYP
 
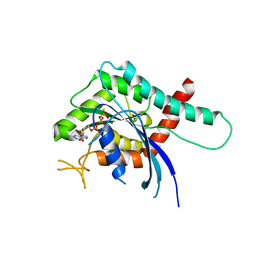 | | GTPase domain of Septin 9 in complex with GTP-gamma-S | | 分子名称: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, Septin-9 | | 著者 | Matos, S.S, Leonardo, D.A, Pereira, H.M, Horjales, E, Araujo, A.P.U, Garratt, R.C. | | 登録日 | 2015-07-30 | | 公開日 | 2016-08-03 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.893 Å) | | 主引用文献 | A complete compendium of crystal structures for the human SEPT3 subgroup reveals functional plasticity at a specific septin interface
Iucrj, 2020
|
|
6KLX
 
 | | Pore structure of Iota toxin binding component (Ib) | | 分子名称: | CALCIUM ION, Iota toxin component Ib | | 著者 | Yoshida, T, Yamada, T, Kawamoto, A, Mitsuoka, K, Iwasaki, K, Tsuge, H. | | 登録日 | 2019-07-30 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Cryo-EM structures reveal translocational unfolding in the clostridial binary iota toxin complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6DKU
 
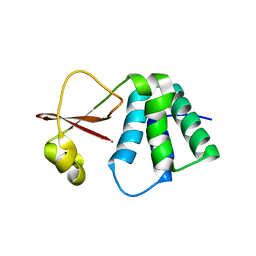 | | Crystal structure of Myotis VP35 mutant of interferon inhibitory domain | | 分子名称: | VP35 | | 著者 | Liu, H, Ginell, G.M, Keefe, L.J, Leung, D.W, Amarasinghe, G.K. | | 登録日 | 2018-05-30 | | 公開日 | 2018-07-18 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Conservation of Structure and Immune Antagonist Functions of Filoviral VP35 Homologs Present in Microbat Genomes.
Cell Rep, 24, 2018
|
|
5CYO
 
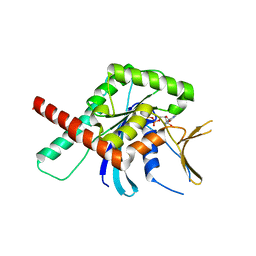 | | High resolution Septin 9 GTPase domain in complex with GDP | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Septin-9 | | 著者 | Matos, S.S, Leonardo, D.A, Pereira, H.M, Horjales, E, Araujo, A.P.U, Garratt, R.C. | | 登録日 | 2015-07-30 | | 公開日 | 2016-08-03 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.0354 Å) | | 主引用文献 | A complete compendium of crystal structures for the human SEPT3 subgroup reveals functional plasticity at a specific septin interface
Iucrj, 2020
|
|
6KLO
 
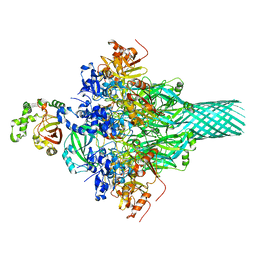 | | Complex structure of Iota toxin enzymatic component (Ia) and binding component (Ib) pore with short stem | | 分子名称: | CALCIUM ION, Iota toxin component Ia, Iota toxin component Ib | | 著者 | Yoshida, T, Yamada, T, Kawamoto, A, Mitsuoka, K, Iwasaki, K, Tsuge, H. | | 登録日 | 2019-07-30 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Cryo-EM structures reveal translocational unfolding in the clostridial binary iota toxin complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6KSD
 
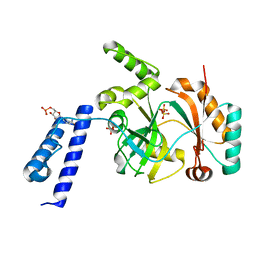 | |
6KRH
 
 | | Structural basis for domain rotation during adenylation of active site K123 and fragment library screening against NAD+ -dependent DNA ligase from Mycobacterium tuberculosis | | 分子名称: | ADENOSINE MONOPHOSPHATE, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase A, ... | | 著者 | Ramachandran, R, Shukla, A, Afsar, M. | | 登録日 | 2019-08-21 | | 公開日 | 2020-08-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Salt bridges at the subdomain interfaces of the adenylation domain and active-site residues of Mycobacterium tuberculosis NAD + -dependent DNA ligase A (MtbLigA) are important for the initial steps of nick-sealing activity.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6KSC
 
 | |
3O40
 
 | | Complex of a chimeric alpha/beta-peptide based on the gp41 CHR domain bound to gp41-5 | | 分子名称: | CHLORIDE ION, GLYCEROL, NONAETHYLENE GLYCOL, ... | | 著者 | Horne, W.S, Johnson, L.M, Gellman, S.H. | | 登録日 | 2010-07-26 | | 公開日 | 2011-07-27 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Broad Distribution of Energetically Important Contacts across an Extended Protein Interface.
J.Am.Chem.Soc., 133, 2011
|
|
3O3Z
 
 | |
1JQ6
 
 | |
3O3X
 
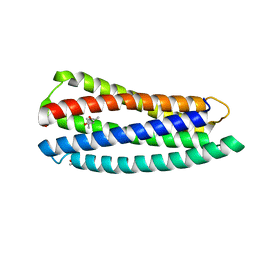 | | Crystal structure of gp41-5, a single-chain 5-helix-bundle based on HIV gp41 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ISOPROPYL ALCOHOL, gp41-5 | | 著者 | Horne, W.S, Johnson, L.M, Gellman, S.H. | | 登録日 | 2010-07-26 | | 公開日 | 2011-07-27 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Broad Distribution of Energetically Important Contacts across an Extended Protein Interface.
J.Am.Chem.Soc., 133, 2011
|
|
4DFC
 
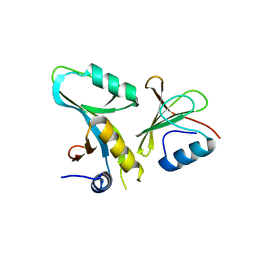 | | Core UvrA/TRCF complex | | 分子名称: | Transcription-repair-coupling factor, UvrABC system protein A | | 著者 | Deaconescu, A.M, Grigorieff, N. | | 登録日 | 2012-01-23 | | 公開日 | 2012-05-02 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.803 Å) | | 主引用文献 | Nucleotide excision repair (NER) machinery recruitment by the transcription-repair coupling factor involves unmasking of a conserved intramolecular interface.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1NZ6
 
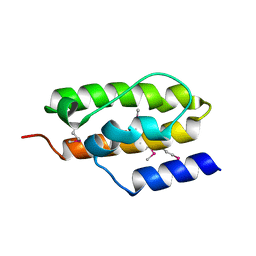 | | Crystal Structure of Auxilin J-Domain | | 分子名称: | Auxilin | | 著者 | Jiang, J, Taylor, A.B, Prasad, K, Ishikawa-Brush, Y, Hart, P.J, Lafer, E.M, Sousa, R. | | 登録日 | 2003-02-16 | | 公開日 | 2003-04-22 | | 最終更新日 | 2011-11-16 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure-function analysis of the auxilin J-domain reveals an extended Hsc70 interaction interface.
Biochemistry, 42, 2003
|
|
7T6F
 
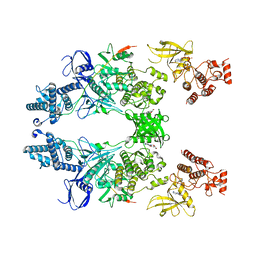 | | Structure of active Janus Kinase (JAK) dimer complexed with cytokine receptor intracellular domain | | 分子名称: | ADENOSINE, ADENOSINE-5'-DIPHOSPHATE, Interferon lambda receptor 1, ... | | 著者 | Glassman, C.R, Tsutsumi, N, Jude, K.M, Garcia, K.C. | | 登録日 | 2021-12-13 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structure of a Janus kinase cytokine receptor complex reveals the basis for dimeric activation.
Science, 376, 2022
|
|
3MI8
 
 | | The structure of TL1A-DCR3 COMPLEX | | 分子名称: | TUMOR NECROSIS FACTOR LIGAND SUPERFAMILY MEMBER 15, SECRETED FORM, Tumor necrosis factor receptor superfamily member 6B | | 著者 | Zhan, C, Patskovsky, Y, Yan, Q, Li, Z, Ramagopal, U.A, Nathenson, S.G, Almo, S.C. | | 登録日 | 2010-04-09 | | 公開日 | 2011-02-23 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.951 Å) | | 主引用文献 | Decoy Strategies: The Structure of TL1A:DcR3 Complex.
Structure, 19, 2011
|
|
3O43
 
 | |
