3I7U
 
 | | Crystal structure of AP4A hydrolase (aq_158) from Aquifex aeolicus VF5 | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AP4A hydrolase, ... | | 著者 | Jeyakanthan, J, Kanaujia, S.P, Nakagawa, N, Sekar, K, Kuramitsu, S, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2009-07-09 | | 公開日 | 2009-07-21 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Free and ATP-bound structures of Ap(4)A hydrolase from Aquifex aeolicus V5
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1SDT
 
 | | Crystal structures of HIV protease V82A and L90M mutants reveal changes in indinavir binding site. | | 分子名称: | CHLORIDE ION, N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE, protease RETROPEPSIN | | 著者 | Mahalingam, B, Wang, Y.-F, Boross, P.I, Tozser, J, Louis, J.M, Harrison, R.W, Weber, I.T. | | 登録日 | 2004-02-14 | | 公開日 | 2004-05-25 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Crystal structures of HIV protease V82A and L90M
mutants reveal changes in the indinavir-binding site
Eur.J.Biochem., 271, 2004
|
|
3ESA
 
 | | cut-1b; NCN-Pt-Pincer-Cutinase Hybrid | | 分子名称: | (2,6-bis[(dimethylamino-kappaN)methyl]-4-{3-[(S)-ethoxy(4-nitrophenoxy)phosphoryl]propyl}phenyl-kappaC~1~)(chloro)platinum(2+), Cutinase 1 | | 著者 | Rutten, L, Mannie, J.P.B.A, Lutz, M, Gros, P. | | 登録日 | 2008-10-05 | | 公開日 | 2009-07-28 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Solid-state structural characterization of cutinase-ECE-pincer-metal hybrids
Chemistry, 15, 2009
|
|
3IEI
 
 | |
1SGT
 
 | |
3QRM
 
 | |
2ZAK
 
 | | Orthorhombic crystal structure of precursor E. coli isoaspartyl peptidase/L-asparaginase (EcAIII) with active-site T179A mutation | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, L-asparaginase precursor, ... | | 著者 | Michalska, K, Hernandez-Santoyo, A, Jaskolski, M. | | 登録日 | 2007-10-07 | | 公開日 | 2008-03-25 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Crystal packing of plant-type L-asparaginase from Escherichia coli
Acta Crystallogr.,Sect.D, 64, 2008
|
|
5RFF
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102704 | | 分子名称: | 1-{4-[(4-chlorophenyl)sulfonyl]piperazin-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | 登録日 | 2020-03-15 | | 公開日 | 2020-03-25 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RFV
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102306 | | 分子名称: | 1-[4-(thiophene-2-carbonyl)piperazin-1-yl]ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | 登録日 | 2020-03-15 | | 公開日 | 2020-03-25 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
2Z76
 
 | | X-ray crystal structure of RV0760c from Mycobacterium tuberculosis at 1.82 Angstrom resolution | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | 著者 | Cherney, M.M, Garen, C.R, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | 登録日 | 2007-08-16 | | 公開日 | 2007-09-18 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Crystal structure of Mycobacterium tuberculosis Rv0760c at 1.50 A resolution, a structural homolog of Delta(5)-3-ketosteroid isomerase.
Biochim.Biophys.Acta, 1784, 2008
|
|
3UU1
 
 | | Anthranilate phosphoribosyltransferase (trpD) from Mycobacterium tuberculosis (complex with inhibitor ACS142) | | 分子名称: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 2-amino-3,5-dimethylbenzoic acid, Anthranilate phosphoribosyltransferase, ... | | 著者 | Castell, A, Short, F.L, Lott, J.S. | | 登録日 | 2011-11-27 | | 公開日 | 2012-12-12 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | The Substrate Capture Mechanism of Mycobacterium tuberculosis Anthranilate Phosphoribosyltransferase Provides a Mode for Inhibition.
Biochemistry, 52, 2013
|
|
5RW6
 
 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z2856434816 | | 分子名称: | 1-(pyridin-4-yl)-N-[(thiophen-2-yl)methyl]methanamine, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | 著者 | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | 登録日 | 2020-10-30 | | 公開日 | 2020-11-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
2ZIS
 
 | | Crystal Structure of rat protein farnesyltransferase complexed with a bezoruran inhibitor and FPP | | 分子名称: | 3-{2-[(S)-(4-cyanophenyl)(hydroxy)(1-methyl-1H-imidazol-5-yl)methyl]-5-nitro-1-benzofuran-7-yl}benzonitrile, ACETIC ACID, FARNESYL DIPHOSPHATE, ... | | 著者 | Fukami, T.A, Sogabe, S, Nagata, Y, Kondoh, O, Ishii, N. | | 登録日 | 2008-02-22 | | 公開日 | 2009-02-24 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Synthesis and structure-activity relationships of novel benzofuran farnesyltransferase inhibitors
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4JIB
 
 | | Crystal structure of of PDE2-inhibitor complex | | 分子名称: | 1-(2-hydroxyethyl)-3-(2-methylbutan-2-yl)-5-[4-(2-methyl-1H-imidazol-1-yl)phenyl]-6,7-dihydropyrazolo[4,3-e][1,4]diazepin-8(1H)-one, MAGNESIUM ION, ZINC ION, ... | | 著者 | Pandit, J. | | 登録日 | 2013-03-05 | | 公開日 | 2013-05-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Discovery of potent, selective, bioavailable phosphodiesterase 2 (PDE2) inhibitors active in an osteoarthritis pain model, Part I: Transformation of selective pyrazolodiazepinone phosphodiesterase 4 (PDE4) inhibitors into selective PDE2 inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4JGV
 
 | | Crystal Structure of Human Nur77 Ligand-binding Domain in Complex with THPN | | 分子名称: | 1-(3,4,5-trihydroxyphenyl)nonan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | 著者 | Zhang, Q, Li, F, Li, A, Tian, X, Wan, W, Wan, Y, Chen, H, Xing, Y, Wu, Q, Lin, T. | | 登録日 | 2013-03-04 | | 公開日 | 2013-12-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.01 Å) | | 主引用文献 | Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway.
Nat.Chem.Biol., 10, 2014
|
|
5RWP
 
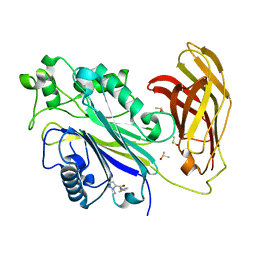 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z915492990 | | 分子名称: | 1-methyl-N-[(thiophen-2-yl)methyl]-1H-pyrazole-5-carboxamide, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | 著者 | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | 登録日 | 2020-10-30 | | 公開日 | 2020-11-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
5RXF
 
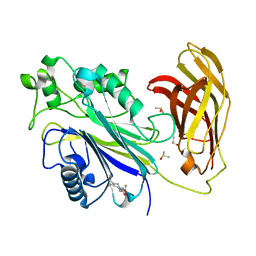 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z2074076908 | | 分子名称: | 1-(5-azaspiro[2.5]octan-5-yl)-2-(difluoromethoxy)ethan-1-one, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | 著者 | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | 登録日 | 2020-10-30 | | 公開日 | 2020-11-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.26 Å) | | 主引用文献 | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
2K4A
 
 | |
5RXX
 
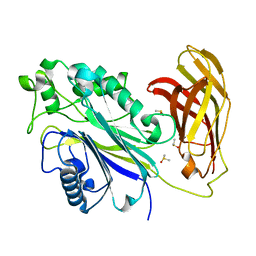 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z1275599911 | | 分子名称: | 1-(2-azaspiro[5.6]dodecan-2-yl)ethan-1-one, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | 著者 | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | 登録日 | 2020-10-30 | | 公開日 | 2020-11-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
1ZOH
 
 | | Crystal structure of protein kinase CK2 in complex with TBB-derivatives inhibitors | | 分子名称: | 1,2-ETHANEDIOL, 5,6,7,8-TETRABROMO-1-METHYL-2,3-DIHYDRO-1H-IMIDAZO[1,2-A]BENZIMIDAZOLE, CHLORIDE ION, ... | | 著者 | Battistutta, R, Mazzorana, M, Sarno, S, Kazimierczuk, Z, Zanotti, G, Pinna, L.A. | | 登録日 | 2005-05-13 | | 公開日 | 2005-11-29 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Inspecting the structure-activity relationship of protein kinase CK2 inhibitors derived from tetrabromo-benzimidazole.
Chem.Biol., 12, 2005
|
|
7G1M
 
 | | Crystal Structure of human FABP4 binding site mutated to that of FABP5 in complex with rac-(1R,2R)-2-[[3-(3-methyl-1,2,4-oxadiazol-5-yl)-4,5,6,7-tetrahydro-1-benzothiophen-2-yl]carbamoyl]cyclohexane-1-carboxylic acid, i.e. SMILES C1(=C(C2=C(S1)CCCC2)C1=NC(=NO1)C)NC(=O)[C@@H]1[C@H](CCCC1)C(=O)O with IC50=0.365 microM | | 分子名称: | (1R,2S)-2-{[(3M)-3-(3-methyl-1,2,4-oxadiazol-5-yl)-4,5,6,7-tetrahydro-1-benzothiophen-2-yl]carbamoyl}cyclohexane-1-carboxylic acid, Fatty acid-binding protein, adipocyte | | 著者 | Ehler, A, Benz, J, Obst, U, Neidhart, W, Rudolph, M.G. | | 登録日 | 2023-04-27 | | 公開日 | 2023-06-14 | | 最終更新日 | 2025-08-13 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
3QK0
 
 | | Crystal structure of PI3K-gamma in complex with benzothiazole 82 | | 分子名称: | N-[6-(6-chloro-5-{[(4-fluorophenyl)sulfonyl]amino}pyridin-3-yl)-1,3-benzothiazol-2-yl]acetamide, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | 著者 | Whittington, D.A, Tang, J, Yakowec, P. | | 登録日 | 2011-01-31 | | 公開日 | 2011-03-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Discovery and Optimization of a Series of Benzothiazole Phosphoinositide 3-Kinase (PI3K)/Mammalian Target of Rapamycin (mTOR) Dual Inhibitors.
J.Med.Chem., 54, 2011
|
|
3QLW
 
 | | Candida albicans dihydrofolate reductase complexed with NADPH and 5-[3-(2,5-dimethoxyphenyl)prop-1-yn-1-yl]-6-ethylpyrimidine-2,4-diamine (UCP120B) | | 分子名称: | 5-[3-(2,5-dimethoxyphenyl)prop-1-yn-1-yl]-6-ethylpyrimidine-2,4-diamine, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative uncharacterized protein CaJ7.0360 | | 著者 | Paulsen, J.L, Bendel, S.D, Anderson, A.C. | | 登録日 | 2011-02-03 | | 公開日 | 2011-07-20 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.504 Å) | | 主引用文献 | Crystal Structures of Candida albicans Dihydrofolate Reductase Bound to Propargyl-Linked Antifolates Reveal the Flexibility of Active Site Loop Residues Critical for Ligand Potency and Selectivity.
Chem.Biol.Drug Des., 78, 2011
|
|
7ZD6
 
 | | Complex I from Ovis aries, at pH7.4, Open state | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | 著者 | Sazanov, L, Petrova, O. | | 登録日 | 2022-03-29 | | 公開日 | 2022-09-21 | | 最終更新日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (3.16 Å) | | 主引用文献 | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
4BAP
 
 | | Hen egg-white lysozyme structure in complex with the europium tris- hydroxyethylcholinetriazoledipicolinate complex at 1.21 A resolution. | | 分子名称: | ACETATE ION, CHLORIDE ION, EUROPIUM (III) ION, ... | | 著者 | Talon, R, Kahn, R, Gautier, A, Nauton, L, Girard, E. | | 登録日 | 2012-09-14 | | 公開日 | 2012-11-14 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.207 Å) | | 主引用文献 | Clicked Europium Dipicolinate Complexes for Protein X-Ray Structure Determination.
Chem.Commun.(Camb.), 48, 2012
|
|
