2XGW
 
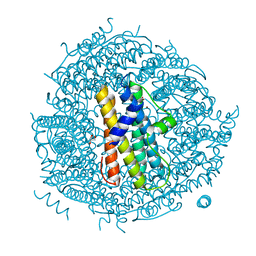 | | ZINC-BOUND CRYSTAL STRUCTURE OF STREPTOCOCCUS PYOGENES DPR | | 分子名称: | CHLORIDE ION, GLYCEROL, PEROXIDE RESISTANCE PROTEIN, ... | | 著者 | Haikarainen, T, Tsou, C.-C, Wu, J.-J, Papageorgiou, A.C. | | 登録日 | 2010-06-08 | | 公開日 | 2010-08-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Characterization and Biological Implications of Di-Zinc Binding in the Ferroxidase Center of Streptococcus Pyogenes Dpr.
Biochem.Biophys.Res.Commun., 398, 2010
|
|
7Y0R
 
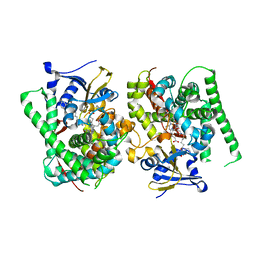 | | Crystal structure of the P450 BM3 heme domain mutant F87L/V78S/A184V in complex with N-imidazolyl-hexanoyl-L-phenylalanine, p-toluidine and hydroxylamine | | 分子名称: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, 4-METHYLANILINE, Bifunctional cytochrome P450/NADPH--P450 reductase, ... | | 著者 | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | 登録日 | 2022-06-06 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Crystal structure of the P450 BM3 heme domain mutant F87L/V78S/A184V in complex with N-imidazolyl-hexanoyl-L-phenylalanine, p-toluidine and hydroxylamine
To Be Published
|
|
2XFY
 
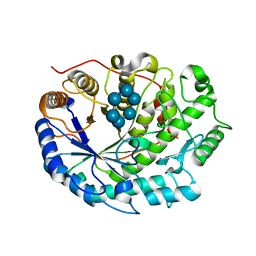 | | Crystal structure of Barley Beta-Amylase complexed with alpha- cyclodextrin | | 分子名称: | 1,2-ETHANEDIOL, BETA-AMYLASE, Cyclohexakis-(1-4)-(alpha-D-glucopyranose) | | 著者 | Rejzek, M, Stevenson, C.E.M, Southard, A.M, Stanley, D, Denyer, K, Smith, A.M, Naldrett, M.J, Lawson, D.M, Field, R.A. | | 登録日 | 2010-05-28 | | 公開日 | 2010-12-01 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.207 Å) | | 主引用文献 | Chemical Genetics and Cereal Starch Metabolism: Structural Basis of the Non-Covalent and Covalent Inhibition of Barley Beta-Amylase.
Mol.Biosyst., 7, 2011
|
|
7Y0K
 
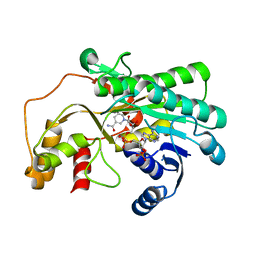 | |
4NG8
 
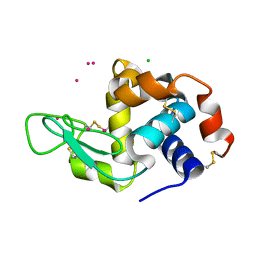 | | Dialyzed HEW lysozyme batch crystallized in 1.9 M CsCl and collected at 100 K. | | 分子名称: | CESIUM ION, CHLORIDE ION, Lysozyme C | | 著者 | Benas, P, Legrand, L, Ries-Kautt, M. | | 登録日 | 2013-11-01 | | 公開日 | 2014-05-28 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.09 Å) | | 主引用文献 | Weak protein-cationic co-ion interactions addressed by X-ray crystallography and mass spectrometry.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7Y0Q
 
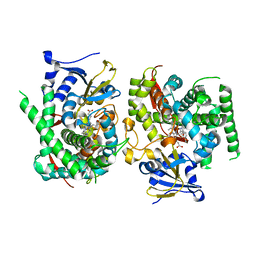 | |
3EXF
 
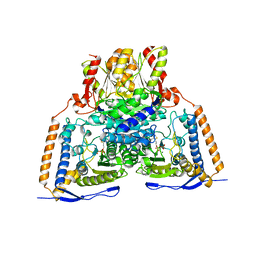 | | Crystal structure of the pyruvate dehydrogenase (E1p) component of human pyruvate dehydrogenase complex | | 分子名称: | MAGNESIUM ION, POTASSIUM ION, Pyruvate dehydrogenase E1 component subunit alpha, ... | | 著者 | Kato, M, Wynn, R.M, Chuang, J.L, Tso, S.-C, Machius, M, Li, J, Chuang, D.T. | | 登録日 | 2008-10-16 | | 公開日 | 2008-11-25 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.998 Å) | | 主引用文献 | Structural basis for inactivation of the human pyruvate dehydrogenase complex by phosphorylation: role of disordered phosphorylation loops.
Structure, 16, 2008
|
|
2GH5
 
 | | Crystal Structure of human Glutathione Reductase complexed with a Fluoro-Analogue of the Menadione Derivative M5 | | 分子名称: | 6-(3-METHYL-1,4-DIOXO-1,4-DIHYDRONAPHTHALEN-2-YL)HEXANOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | 著者 | Fritz-Wolf, K, Winzer, A, Bauer, H, Schirmer, H, Davioud-Charvet, E. | | 登録日 | 2006-03-25 | | 公開日 | 2006-09-26 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | A fluoro analogue of the menadione derivative 6-[2'-(3'-methyl)-1',4'-naphthoquinolyl]hexanoic acid is a suicide substrate of glutathione reductase. Crystal structure of the alkylated human enzyme
J.Am.Chem.Soc., 128, 2006
|
|
4U14
 
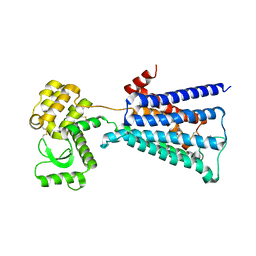 | | Structure of the M3 muscarinic acetylcholine receptor bound to the antagonist tiotropium crystallized with disulfide-stabilized T4 lysozyme (dsT4L) | | 分子名称: | (1R,2R,4S,5S,7S)-7-{[hydroxy(dithiophen-2-yl)acetyl]oxy}-9,9-dimethyl-3-oxa-9-azoniatricyclo[3.3.1.0~2,4~]nonane, Muscarinic acetylcholine receptor M3,Endolysin,Muscarinic acetylcholine receptor M3 | | 著者 | Thorsen, T.S, Matt, R.A, Weis, W.I, Kobilka, B.K. | | 登録日 | 2014-07-15 | | 公開日 | 2014-11-26 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.57 Å) | | 主引用文献 | Modified T4 Lysozyme Fusion Proteins Facilitate G Protein-Coupled Receptor Crystallogenesis.
Structure, 22, 2014
|
|
3L4S
 
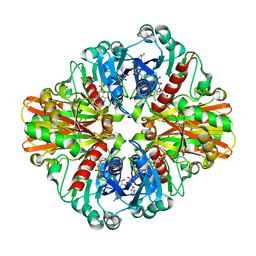 | | Crystal structure of C151G mutant of Glyceraldehyde 3-phosphate dehydrogenase 1 (GAPDH1) from methicillin resistant Staphylococcus aureus MRSA252 complexed with NAD and G3P | | 分子名称: | 3-PHOSPHOGLYCERIC ACID, Glyceraldehyde-3-phosphate dehydrogenase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | 登録日 | 2009-12-21 | | 公開日 | 2010-08-18 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
2R1V
 
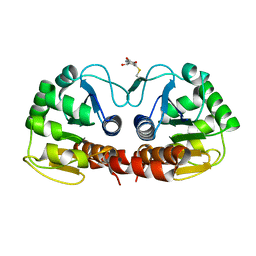 | |
4TWZ
 
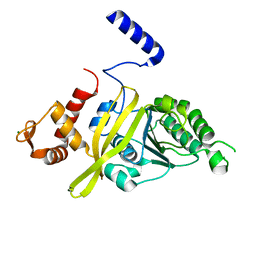 | | Crystal Structure Analysis of E Coli. RecA Protein | | 分子名称: | MAGNESIUM ION, Protein RecA | | 著者 | Hikima, T, Hiraki, T, Furuse, M, Ikawa, S, Iwasaki, W, Shibata, T, Kamiya, N. | | 登録日 | 2014-07-02 | | 公開日 | 2015-07-08 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Loop L1 governs the DNA-binding specificity and order for RecA-catalyzed reactions in homologous recombination and DNA repair
Nucleic Acids Res., 43, 2015
|
|
3EXD
 
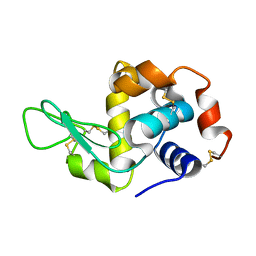 | |
3KNG
 
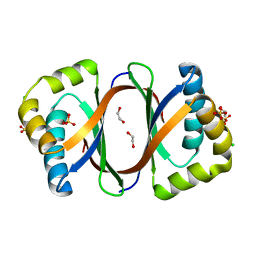 | | Crystal structure of SnoaB, a cofactor-independent oxygenase from Streptomyces nogalater, determined to 1.9 resolution | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | 著者 | Koskiniemi, H, Grocholski, T, Lindqvist, Y, Mantsala, P, Niemi, J, Schneider, G. | | 登録日 | 2009-11-12 | | 公開日 | 2010-01-26 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of the cofactor-independent monooxygenase SnoaB from Streptomyces nogalater: implications for the reaction mechanism
Biochemistry, 49, 2010
|
|
7Y0P
 
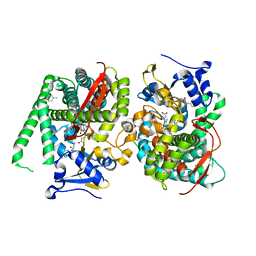 | | Crystal structure of the P450 BM3 heme domain mutant F87A/T268V/A82T/I263L in complex with N-imidazolyl-hexanoyl-L-phenylalanine, p-cresol and hydroxylamine | | 分子名称: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | 著者 | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | 登録日 | 2022-06-06 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Crystal structure of the P450 BM3 heme domain mutant F87A/T268V/A82T/I263L in complex with N-imidazolyl-hexanoyl-L-phenylalanine, p-cresol and hydroxylamine
To Be Published
|
|
3KNT
 
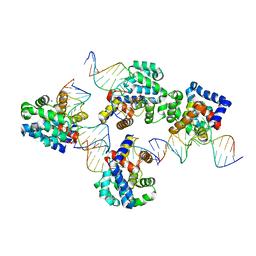 | |
3KOU
 
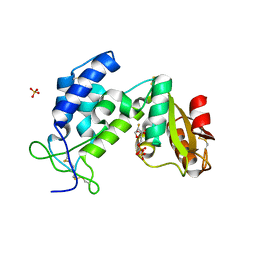 | | Structural insights into the catalytic mechanism of CD38: Evidence for a conformationally flexible covalent enzyme-substrate complex. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD38 molecule, ... | | 著者 | Egea, P.F, Muller-Steffner, H, Stroud, R.M, Oppenheimer, N.J, Kellenberger, E, Schuber, F. | | 登録日 | 2009-11-13 | | 公開日 | 2010-10-27 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Insights into the mechanism of bovine CD38/NAD+glycohydrolase from the X-ray structures of its Michaelis complex and covalently-trapped intermediates.
Plos One, 7, 2012
|
|
3F10
 
 | |
4MOM
 
 | | Pyranose 2-oxidase H450G mutant with 3-fluorinated galactose | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-deoxy-3-fluoro-beta-D-galactopyranose, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | 著者 | Tan, T.C, Spadiut, O, Gandini, R, Haltrich, D, Divne, C. | | 登録日 | 2013-09-12 | | 公開日 | 2014-02-05 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural Basis for Binding of Fluorinated Glucose and Galactose to Trametes multicolor Pyranose 2-Oxidase Variants with Improved Galactose Conversion.
Plos One, 9, 2014
|
|
7Y0T
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-phenylalanyl-L-phenylalanine | | 分子名称: | Bifunctional cytochrome P450/NADPH--P450 reductase, I7X-PHE-PHE, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | 登録日 | 2022-06-06 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Anchoring a Structurally Editable Proximal Cofactor-like Module to Construct an Artificial Dual-center Peroxygenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
3EXG
 
 | | Crystal structure of the pyruvate dehydrogenase (E1p) component of human pyruvate dehydrogenase complex | | 分子名称: | POTASSIUM ION, Pyruvate dehydrogenase E1 component subunit alpha, somatic form, ... | | 著者 | Kato, M, Wynn, R.M, Chuang, J.L, Tso, S.-C, Machius, M, Li, J, Chuang, D.T. | | 登録日 | 2008-10-16 | | 公開日 | 2008-11-25 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.011 Å) | | 主引用文献 | Structural basis for inactivation of the human pyruvate dehydrogenase complex by phosphorylation: role of disordered phosphorylation loops.
Structure, 16, 2008
|
|
2QCK
 
 | |
2GD1
 
 | |
3KL5
 
 | |
3EZ8
 
 | | Crystal Structure of endoglucanase Cel9A from the thermoacidophilic Alicyclobacillus acidocaldarius | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Cellulase, ... | | 著者 | Pereira, J.H, Sapra, R, Simmons, B, Adams, P.D. | | 登録日 | 2008-10-22 | | 公開日 | 2009-08-04 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.302 Å) | | 主引用文献 | Structure of endoglucanase Cel9A from the thermoacidophilic Alicyclobacillus acidocaldarius
Acta Crystallogr.,Sect.D, 65, 2009
|
|
