1U5S
 
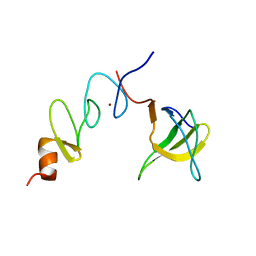 | | NMR structure of the complex between Nck-2 SH3 domain and PINCH-1 LIM4 domain | | 分子名称: | Cytoplasmic protein NCK2, PINCH protein, ZINC ION | | 著者 | Vaynberg, J, Fukuda, T, Vinogradova, O, Velyvis, A, Ng, L, Wu, C, Qin, J. | | 登録日 | 2004-07-28 | | 公開日 | 2005-04-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of an ultraweak protein-protein complex and its crucial role in regulation of cell morphology and motility.
Mol.Cell, 17, 2005
|
|
1R8U
 
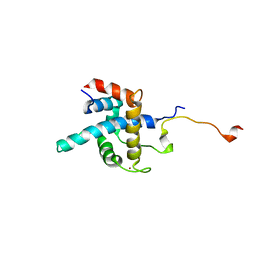 | | NMR structure of CBP TAZ1/CITED2 complex | | 分子名称: | CREB-binding protein, Cbp/p300-interacting transactivator 2, ZINC ION | | 著者 | De Guzman, R.N, Martinez-Yamout, M, Dyson, H.J, Wright, P.E. | | 登録日 | 2003-10-28 | | 公開日 | 2004-03-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Interaction of the TAZ1 domain of the CREB-binding protein with the activation domain of CITED2: regulation by competition between intrinsically unstructured ligands for non-identical binding sites.
J.Biol.Chem., 279, 2004
|
|
6B34
 
 | |
1RKL
 
 | |
6B35
 
 | |
1SV1
 
 | |
1RML
 
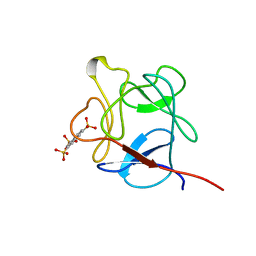 | | NMR STUDY OF ACID FIBROBLAST GROWTH FACTOR BOUND TO 1,3,6-NAPHTHALENE TRISULPHONATE, 26 STRUCTURES | | 分子名称: | ACIDIC FIBROBLAST GROWTH FACTOR, NAPHTHALENE TRISULFONATE | | 著者 | Lozano, R.M, Jimenez, M.A, Santoro, J, Rico, M, Gimenez-Gallego, G. | | 登録日 | 1998-05-21 | | 公開日 | 1998-11-11 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of acidic fibroblast growth factor bound to 1,3, 6-naphthalenetrisulfonate: a minimal model for the anti-tumoral action of suramins and suradistas.
J.Mol.Biol., 281, 1998
|
|
1L1W
 
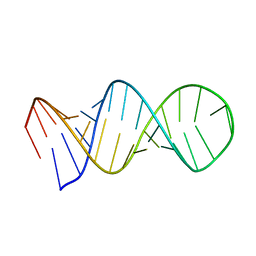 | | NMR structure of a SRP19 binding domain in human SRP RNA | | 分子名称: | SRP19 binding domain of SRP RNA | | 著者 | Sakamoto, T, Morita, S, Tabata, K, Nakamura, K, Kawai, G. | | 登録日 | 2002-02-20 | | 公開日 | 2002-05-22 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a SRP19 binding domain in human SRP RNA.
J.Biochem.(Tokyo), 132, 2002
|
|
1LFU
 
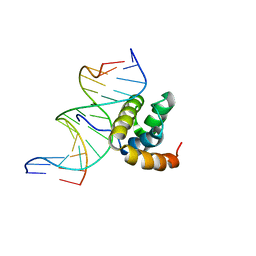 | | NMR Solution Structure of the Extended PBX Homeodomain Bound to DNA | | 分子名称: | 5'-D(*GP*CP*GP*CP*AP*TP*GP*AP*TP*TP*GP*CP*CP*C)-3', 5'-D(*GP*GP*GP*CP*AP*AP*TP*CP*AP*TP*GP*CP*GP*C)-3', homeobox protein PBX1 | | 著者 | Sprules, T, Green, N, Featherstone, M, Gehring, K. | | 登録日 | 2002-04-12 | | 公開日 | 2003-01-14 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Lock and Key Binding of the HOX YPWM Peptide to the PBX Homeodomain
J.Biol.Chem., 278, 2003
|
|
1BAU
 
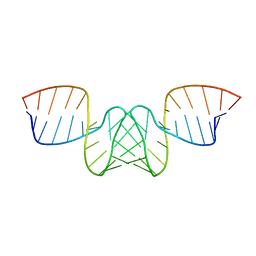 | | NMR STRUCTURE OF THE DIMER INITIATION COMPLEX OF HIV-1 GENOMIC RNA, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | SL1 RNA DIMER | | 著者 | Mujeeb, A, Clever, J.L, Billeci, T.M, James, T.L, Parslow, T.G. | | 登録日 | 1998-04-18 | | 公開日 | 1999-04-27 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the dimer initiation complex of HIV-1 genomic RNA.
Nat.Struct.Biol., 5, 1998
|
|
2MI0
 
 | |
1I6G
 
 | |
1I6F
 
 | |
1SG7
 
 | | NMR solution structure of the putative cation transport regulator ChaB | | 分子名称: | Putative Cation transport regulator chaB | | 著者 | Osborne, M.J, Siddiqui, N, Cygler, M, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | 登録日 | 2004-02-23 | | 公開日 | 2005-03-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of ChaB, a putative membrane ion antiporter regulator from Escherichia coli
BMC STRUCT.BIOL., 4, 2004
|
|
1TWO
 
 | |
1BLJ
 
 | | NMR ENSEMBLE OF BLK SH2 DOMAIN, 20 STRUCTURES | | 分子名称: | P55 BLK PROTEIN TYROSINE KINASE | | 著者 | Metzler, W.J, Leiting, B, Pryor, K, Mueller, L, Farmer II, B.T. | | 登録日 | 1996-03-26 | | 公開日 | 1997-03-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The three-dimensional solution structure of the SH2 domain from p55blk kinase.
Biochemistry, 35, 1996
|
|
2MTK
 
 | |
1APJ
 
 | | NMR STUDY OF THE TRANSFORMING GROWTH FACTOR BETA BINDING PROTEIN-LIKE DOMAIN (TB MODULE/8-CYS DOMAIN), NMR, 21 STRUCTURES | | 分子名称: | FIBRILLIN | | 著者 | Yuan, X, Downing, A.K, Knott, V, Handford, P.A. | | 登録日 | 1997-07-22 | | 公開日 | 1998-01-28 | | 最終更新日 | 2021-11-03 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the transforming growth factor beta-binding protein-like module, a domain associated with matrix fibrils.
EMBO J., 16, 1997
|
|
1BLK
 
 | | NMR ENSEMBLE OF BLK SH2 DOMAIN USING CHEMICAL SHIFT REFINEMENT, 20 STRUCTURES | | 分子名称: | P55 BLK PROTEIN TYROSINE KINASE | | 著者 | Metzler, W.J, Leiting, B, Pryor, K, Mueller, L, Farmer II, B.T. | | 登録日 | 1996-03-26 | | 公開日 | 1997-03-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The three-dimensional solution structure of the SH2 domain from p55blk kinase.
Biochemistry, 35, 1996
|
|
1CQ5
 
 | | NMR STRUCTURE OF SRP RNA DOMAIN IV | | 分子名称: | SRP RNA DOMAIN IV | | 著者 | Schmitz, U, James, T.L, Behrens, S, Freymann, D.M, Lukavsky, P, Walter, P. | | 登録日 | 1999-08-05 | | 公開日 | 1999-08-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the phylogenetically most conserved domain of SRP RNA.
RNA, 5, 1999
|
|
1CMR
 
 | | NMR SOLUTION STRUCTURE OF A CHIMERIC PROTEIN, DESIGNED BY TRANSFERRING A FUNCTIONAL SNAKE BETA-HAIRPIN INTO A SCORPION ALPHA/BETA SCAFFOLD (PH 3.5, 20C), NMR, 18 STRUCTURES | | 分子名称: | CHARYBDOTOXIN, ALPHA CHIMERA | | 著者 | Zinn-Justin, S, Guenneugues, M, Drakopoulou, E, Gilquin, B, Vita, C, Menez, A. | | 登録日 | 1996-03-15 | | 公開日 | 1996-08-01 | | 最終更新日 | 2021-11-03 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Transfer of a beta-hairpin from the functional site of snake curaremimetic toxins to the alpha/beta scaffold of scorpion toxins: three-dimensional solution structure of the chimeric protein.
Biochemistry, 35, 1996
|
|
2MTJ
 
 | |
2M6T
 
 | | NMR solution structure ensemble of 3-4D mutant domain 11 IGF2R | | 分子名称: | Insulin-like growth factor 2 receptor variant | | 著者 | Strickland, M, Williams, C, Richards, E, Minnall, L, Crump, M.P, Frago, S, Hughes, J, Garner, L, Hoppe, H, Rezgui, D, Zaccheo, O.J, Prince, S.N, Hassan, A.B, Whittaker, S. | | 登録日 | 2013-04-09 | | 公開日 | 2014-10-15 | | 最終更新日 | 2016-06-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Functional evolution of IGF2:IGF2R domain 11 binding generates novel structural interactions and a specific IGF2 antagonist.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1N0K
 
 | | NMR Structure of duplex DNA d(CCAAGGXCTTGGG), X is a 3' phosphoglycolate, 5'phosphate gapped lesion | | 分子名称: | 2-PHOSPHOGLYCOLIC ACID, 5'-D(*CP*CP*AP*AP*GP*G)-3', 5'-D(*CP*CP*CP*AP*AP*GP*GP*CP*CP*TP*TP*GP*G)-3', ... | | 著者 | Junker, H.-D, Hoehn, S.T, Bunt, R.C, Marathius, V, Chen, J, Turner, C.J, Stubbe, J. | | 登録日 | 2002-10-14 | | 公開日 | 2003-01-07 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Synthesis, Characterization and Solution Structure of Tethered Oligonucleotides Containing an Internal 3'-Phosphoglycolate, 5'-Phosphate Gapped Lesion
Nucleic Acids Res., 30, 2002
|
|
1GIZ
 
 | |
