7ANW
 
 | | hSARM1 NAD+ complex | | 分子名称: | NAD(+) hydrolase SARM1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Sporny, M, Guez-Haddad, J, Khazma, T, Yaron, A, Mim, C, Isupov, M.N, Zalk, R, Dessau, M, Hons, M, Opatowsky, Y. | | 登録日 | 2020-10-13 | | 公開日 | 2020-11-11 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.68 Å) | | 主引用文献 | Structural basis for SARM1 inhibition and activation under energetic stress.
Elife, 9, 2020
|
|
7AO8
 
 | | Structure of the MTA1/HDAC1/MBD2 NURD deacetylase complex | | 分子名称: | Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, Metastasis-associated protein MTA1, ... | | 著者 | Millard, C.J, Fairall, L, Ragan, T.J, Savva, C.G, Schwabe, J.W.R. | | 登録日 | 2020-10-14 | | 公開日 | 2020-11-11 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | The topology of chromatin-binding domains in the NuRD deacetylase complex.
Nucleic Acids Res., 48, 2020
|
|
2J3Z
 
 | | Crystal structure of the enzymatic component C2-I of the C2-toxin from Clostridium botulinum at pH 6.1 | | 分子名称: | C2 TOXIN COMPONENT I, COBALT (II) ION, GLYCEROL, ... | | 著者 | Schleberger, C, Hochmann, H, Barth, H, Aktories, K, Schulz, G.E. | | 登録日 | 2006-08-23 | | 公開日 | 2006-10-11 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure and Action of the Binary C2 Toxin from Clostridium Botulinum.
J.Mol.Biol., 364, 2006
|
|
2LZQ
 
 | |
7SV1
 
 | |
3IKJ
 
 | | Structural characterization for the nucleotide binding ability of subunit A mutant S238A of the A1AO ATP synthase | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, V-type ATP synthase alpha chain | | 著者 | Kumar, A, Manimekali, M.S.S, Balakrishna, A.M, Jeyakanthan, J, Gruber, G. | | 登録日 | 2009-08-06 | | 公開日 | 2010-01-12 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Nucleotide binding states of subunit A of the A-ATP synthase and the implication of P-loop switch in evolution.
J.Mol.Biol., 396, 2010
|
|
2ICE
 
 | | CRIg bound to C3c | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Complement C3 alpha chain, ... | | 著者 | Wiesmann, C. | | 登録日 | 2006-09-12 | | 公開日 | 2006-11-07 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structure of C3b in complex with CRIg gives insights into regulation of complement activation.
Nature, 444, 2006
|
|
5KMY
 
 | | Crystal structure of tryptophan synthase subunit alpha from Legionella pneumophila str. Paris | | 分子名称: | Tryptophan synthase alpha chain | | 著者 | Nocek, B, Hatzos-Skintges, C, Endres, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-06-27 | | 公開日 | 2016-08-03 | | 最終更新日 | 2017-02-08 | | 実験手法 | X-RAY DIFFRACTION (1.908 Å) | | 主引用文献 | Crystal structure of tryptophan synthase subunit alpha from Legionella pneumophila str. Paris
To Be Published
|
|
7KRM
 
 | |
3IKY
 
 | | Structural model of ParM filament in the open state by cryo-EM | | 分子名称: | Plasmid segregation protein parM | | 著者 | Galkin, V.E, Orlova, A, Rivera, C, Mullins, R.D, Egelman, E.H. | | 登録日 | 2009-08-06 | | 公開日 | 2009-09-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (18 Å) | | 主引用文献 | Structural polymorphism of the ParM filament and dynamic instability
Structure, 17, 2009
|
|
2ZWZ
 
 | | alpha-L-fucosidase complexed with inhibitor, Core1 | | 分子名称: | (2R,3R,4R,5R,6S)-2-(aminomethyl)-6-methylpiperidine-3,4,5-triol, Alpha-L-fucosidase, putative | | 著者 | Wu, H.-J, Ko, T.-P, Ho, C.-W, Lin, C.-H, Wang, A.H.-J. | | 登録日 | 2008-12-19 | | 公開日 | 2009-12-08 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | Structural basis of alpha-fucosidase inhibition by iminocyclitols with K(i) values in the micro- to picomolar range.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
7ARY
 
 | | Twist-Tower_twist-corrected-variant | | 分子名称: | SCAFFOLD STRAND, STAPLE STRAND | | 著者 | Kube, M, Kohler, F, Feigl, E, Nagel-Yuksel, B, Willner, E.M, Funke, J.J, Gerling, T, Stommer, P, Honemann, M.N, Martin, T.G, Scheres, S.H.W, Dietz, H. | | 登録日 | 2020-10-26 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (8.5 Å) | | 主引用文献 | Revealing the structures of megadalton-scale DNA complexes with nucleotide resolution.
Nat Commun, 11, 2020
|
|
7KUS
 
 | | Crystal Structure of Danio rerio Histone Deacetylase 10 H137A Mutant in Complex with N8-Acetylspermidine (Tetrahedral Intermediate) | | 分子名称: | 1,2-ETHANEDIOL, 1-({4-[(3-aminopropyl)amino]butyl}amino)ethane-1,1-diol, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Herbst-Gervasoni, C.J, Christianson, D.W. | | 登録日 | 2020-11-25 | | 公開日 | 2021-02-03 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | X-ray Crystallographic Snapshots of Substrate Binding in the Active Site of Histone Deacetylase 10.
Biochemistry, 60, 2021
|
|
7TDO
 
 | | Cryo-EM structure of transmembrane AAA+ protease FtsH in the ADP state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent zinc metalloprotease FtsH | | 著者 | Liu, W, Schoonen, M, Wang, T, McSweeney, S, Liu, Q. | | 登録日 | 2022-01-02 | | 公開日 | 2022-04-06 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (3.15 Å) | | 主引用文献 | Cryo-EM structure of transmembrane AAA+ protease FtsH in the ADP state.
Commun Biol, 5, 2022
|
|
2ZXD
 
 | | alpha-L-fucosidase complexed with inhibitor, iso-6FNJ | | 分子名称: | (2S,3R,4S,5R)-2-(1-methylethyl)piperidine-3,4,5-triol, Alpha-L-fucosidase, putative | | 著者 | Wu, H.-J, Ko, T.-P, Ho, C.-W, Lin, C.-H, Wang, A.H.-J. | | 登録日 | 2008-12-22 | | 公開日 | 2009-12-08 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structural basis of alpha-fucosidase inhibition by iminocyclitols with K(i) values in the micro- to picomolar range.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
7SNE
 
 | | Pertussis toxin S1 subunit bound to BaAD | | 分子名称: | Pertussis toxin subunit 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl [(2R,3S,4R,5S)-5-(3-carbamoylanilino)-3,4-dihydroxyoxolan-2-yl]methyl dihydrogen diphosphate (non-preferred name) | | 著者 | Littler, D.R, Beddoe, T. | | 登録日 | 2021-10-28 | | 公開日 | 2022-04-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.00011 Å) | | 主引用文献 | Crystal structures of pertussis toxin with NAD + and analogs provide structural insights into the mechanism of its cytosolic ADP-ribosylation activity.
J.Biol.Chem., 298, 2022
|
|
2ZXY
 
 | | Crystal Structure of Cytochrome c555 from Aquifex aeolicus | | 分子名称: | Cytochrome c552, HEME C | | 著者 | Obuchi, M, Kawahara, K, Motooka, D, Nakamura, S, Yamanaka, M, Takeda, T, Uchiyama, S, Kobayashi, Y, Ohkubo, T, Sambongi, Y. | | 登録日 | 2009-01-09 | | 公開日 | 2009-08-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Hyperstability and crystal structure of cytochrome c(555) from hyperthermophilic Aquifex aeolicus
Acta Crystallogr.,Sect.D, 65, 2009
|
|
7SP8
 
 | | Chlorella virus Hyaluronan Synthase bound to UDP-GlcNAc | | 分子名称: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CHOLESTEROL HEMISUCCINATE, Hyaluronan synthase, ... | | 著者 | Maloney, F.P, Kuklewicz, J, Zimmer, J. | | 登録日 | 2021-11-02 | | 公開日 | 2022-04-06 | | 最終更新日 | 2022-04-20 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structure, substrate recognition and initiation of hyaluronan synthase.
Nature, 604, 2022
|
|
7E5E
 
 | | Crystal structure of GDP-bound GNAS in complex with the cyclic peptide inhibitor GD20 | | 分子名称: | CHLORIDE ION, GD20, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Hu, Q, Dai, S, Shokat, K.M. | | 登録日 | 2021-02-18 | | 公開日 | 2022-03-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | State-selective modulation of heterotrimeric G alpha s signaling with macrocyclic peptides.
Cell, 185, 2022
|
|
3AC0
 
 | | Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose | | 分子名称: | Beta-glucosidase I, beta-D-glucopyranose | | 著者 | Yoshida, E, Hidaka, M, Fushinobu, S, Katayama, T, Kumagai, H. | | 登録日 | 2009-12-25 | | 公開日 | 2010-08-11 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | Role of a PA14 domain in determining substrate specificity of a glycoside hydrolase family 3 beta-glucosidase from Kluyveromyces marxianus.
Biochem.J., 431, 2010
|
|
7SP7
 
 | | Chlorella virus hyaluronan synthase inhibited by UDP | | 分子名称: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CHOLESTEROL HEMISUCCINATE, Hyaluronan synthase, ... | | 著者 | Maloney, F.P, Kuklewicz, J, Zimmer, J. | | 登録日 | 2021-11-02 | | 公開日 | 2022-04-06 | | 最終更新日 | 2022-04-20 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structure, substrate recognition and initiation of hyaluronan synthase.
Nature, 604, 2022
|
|
2ZYI
 
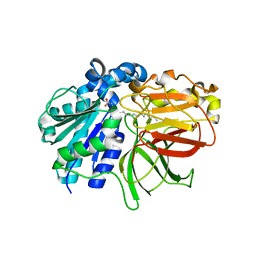 | | A. Fulgidus lipase with fatty acid fragment and calcium | | 分子名称: | CALCIUM ION, Lipase, putative, ... | | 著者 | Chen, C.K, Ko, T.P, Guo, R.T, Wang, A.H. | | 登録日 | 2009-01-22 | | 公開日 | 2009-06-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of the alkalohyperthermophilic Archaeoglobus fulgidus lipase contains a unique C-terminal domain essential for long-chain substrate binding.
J.Mol.Biol., 390, 2009
|
|
2MHR
 
 | |
7SUY
 
 | |
7E73
 
 | |
