6JMP
 
 | |
6JR8
 
 | | Flavobacterium johnsoniae GH31 dextranase, FjDex31A, mutant D412A complexed with isomaltotriose | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Candidate alpha-glycosidase Glycoside hydrolase family 31, ... | | 著者 | Tonozuka, T. | | 登録日 | 2019-04-02 | | 公開日 | 2019-04-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural insights into polysaccharide recognition by Flavobacterium johnsoniae dextranase, a member of glycoside hydrolase family 31.
Febs J., 287, 2020
|
|
7VT9
 
 | |
5Z34
 
 | |
2HE5
 
 | | Crystal structure of 17alpha-hydroxysteroid dehydrogenase in binary complex with NADP(H) in an open conformation | | 分子名称: | ACETATE ION, Aldo-keto reductase family 1, member C21, ... | | 著者 | Faucher, F, Pereira de Jesus-Tran, K, Cantin, L, Luu-the, V, Labrie, F, Breton, R. | | 登録日 | 2006-06-21 | | 公開日 | 2006-12-05 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal Structures of Mouse 17alpha-Hydroxysteroid Dehydrogenase (Apoenzyme and Enzyme-NADP(H) Binary Complex): Identification of Molecular Determinants Responsible for the Unique 17alpha-reductive Activity of this Enzyme.
J.Mol.Biol., 364, 2006
|
|
5Z3B
 
 | | Glycosidase Y48F | | 分子名称: | CITRIC ACID, GLYCEROL, Glycoside hydrolase 15-related protein | | 著者 | Tanaka, Y, Chen, M, Tagami, T, Yao, M, Kimura, A. | | 登録日 | 2018-01-05 | | 公開日 | 2019-05-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
2HEA
 
 | | CONTRIBUTION OF WATER MOLECULES IN THE INTERIOR OF A PROTEIN TO THE CONFORMATIONAL STABILITY | | 分子名称: | LYSOZYME, SODIUM ION | | 著者 | Takano, K, Funahashi, J, Yamagata, Y, Fujii, S, Yutani, K. | | 登録日 | 1997-09-16 | | 公開日 | 1998-01-14 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Contribution of water molecules in the interior of a protein to the conformational stability.
J.Mol.Biol., 274, 1997
|
|
6JND
 
 | | Cryo-EM structure of glutamate dehydrogenase from Thermococcus profundus | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glutamate dehydrogenase | | 著者 | Oide, M, Kato, T, Oroguchi, T, Nakasako, M. | | 登録日 | 2019-03-14 | | 公開日 | 2020-02-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Energy landscape of domain motion in glutamate dehydrogenase deduced from cryo-electron microscopy.
Febs J., 287, 2020
|
|
6COX
 
 | |
2HEF
 
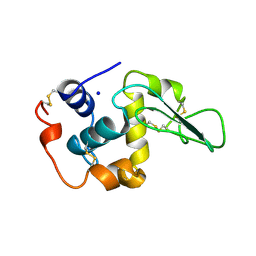 | | CONTRIBUTION OF WATER MOLECULES IN THE INTERIOR OF A PROTEIN TO THE CONFORMATIONAL STABILITY | | 分子名称: | LYSOZYME, SODIUM ION | | 著者 | Takano, K, Funahashi, J, Yamagata, Y, Fujii, S, Yutani, K. | | 登録日 | 1997-09-16 | | 公開日 | 1998-01-14 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Contribution of water molecules in the interior of a protein to the conformational stability.
J.Mol.Biol., 274, 1997
|
|
6JNN
 
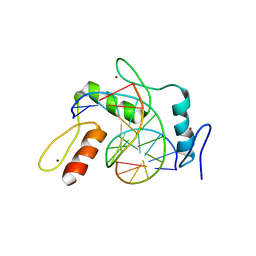 | | REF6 ZnF2-4-NAC004-mC1 complex | | 分子名称: | DNA (5'-D(*CP*AP*AP*AP*AP*CP*AP*GP*AP*GP*AP*A)-3'), DNA (5'-D(*TP*TP*(5CM)P*TP*CP*TP*GP*TP*TP*TP*TP*G)-3'), Lysine-specific demethylase REF6, ... | | 著者 | Yao, Q.Q, Wu, B.X, Ma, J.B. | | 登録日 | 2019-03-17 | | 公開日 | 2019-03-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | DNA methylation repels targeting of Arabidopsis REF6.
Nat Commun, 10, 2019
|
|
3DGH
 
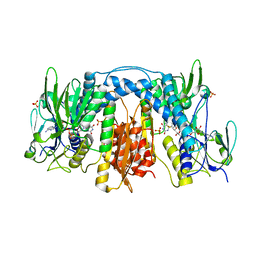 | | Crystal Structure of Drosophila Thioredoxin Reductase, C-terminal 8-residue truncation | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, Thioredoxin reductase 1, ... | | 著者 | Eckenroth, B.E, Hondal, R.J, Everse, S.J. | | 登録日 | 2008-06-13 | | 公開日 | 2009-06-16 | | 最終更新日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (1.745 Å) | | 主引用文献 | Crystal Structure of Drosophila Thioredoxin Reductase, C-terminal 8-residue truncation
To be Published
|
|
6CQ9
 
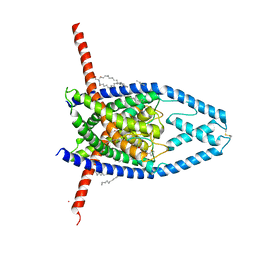 | | K2P2.1(TREK-1):ML402 complex | | 分子名称: | CADMIUM ION, HEXADECANE, N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)PALMITAMIDE, ... | | 著者 | Lolicato, M, Minor, D.L. | | 登録日 | 2018-03-14 | | 公開日 | 2018-04-11 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | K2P2.1 (TREK-1)-activator complexes reveal a cryptic selectivity filter binding site.
Nature, 547, 2017
|
|
7VR2
 
 | |
3DHU
 
 | | Crystal structure of an alpha-amylase from Lactobacillus plantarum | | 分子名称: | Alpha-amylase | | 著者 | Bonanno, J.B, Dickey, M, Bain, K.T, Iizuka, M, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-06-18 | | 公開日 | 2008-08-12 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of an alpha-amylase from Lactobacillus plantarum
To be Published
|
|
7VR4
 
 | |
2WYE
 
 | | The quorum quenching N-acyl homoserine lactone acylase PvdQ is an Ntn- Hydrolase with an unusual substrate-binding pocket | | 分子名称: | ACYL-HOMOSERINE LACTONE ACYLASE PVDQ SUBUNIT ALPHA, ACYL-HOMOSERINE LACTONE ACYLASE PVDQ SUBUNIT BETA, GLYCEROL | | 著者 | Bokhove, M, Nadal Jimenez, P, Quax, W.J, Dijkstra, B.W. | | 登録日 | 2009-11-16 | | 公開日 | 2009-12-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The Quorum-Quenching N-Acyl Homoserine Lactone Acylase Pvdq is an Ntn-Hydrolase with an Unusual Substrate-Binding Pocket
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6CQ6
 
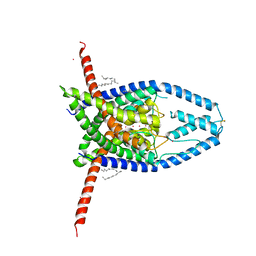 | | K2P2.1(TREK-1) apo structure | | 分子名称: | CADMIUM ION, DECANE, POTASSIUM ION, ... | | 著者 | Lolicato, M, Minor, D.L. | | 登録日 | 2018-03-14 | | 公開日 | 2018-03-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | K2P2.1 (TREK-1)-activator complexes reveal a cryptic selectivity filter binding site.
Nature, 547, 2017
|
|
6EBS
 
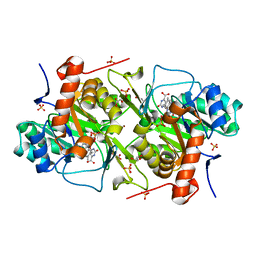 | | Crystal structure of Leishmania major dihydroorotate dehydrogenase mutant H174A in complex with orotate | | 分子名称: | Dihydroorotate dehydrogenase (fumarate), FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | 著者 | Reis, R.A.G, Pinheiro, M.P, de Souza, A.L, Hunter, W.N, Nonato, M.C. | | 登録日 | 2018-08-07 | | 公開日 | 2019-08-21 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Crystal structure of Leishmania major dihydroorotate dehydrogenase mutant H174A in complex with orotate
To Be Published
|
|
4RPE
 
 | |
5Z7N
 
 | | SmChiA sliding-intermediate with chitopentaose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Chitinase A, ... | | 著者 | Nakamura, A, Iino, R. | | 登録日 | 2018-01-30 | | 公開日 | 2018-09-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Processive chitinase is Brownian monorail operated by fast catalysis after peeling rail from crystalline chitin.
Nat Commun, 9, 2018
|
|
3DMT
 
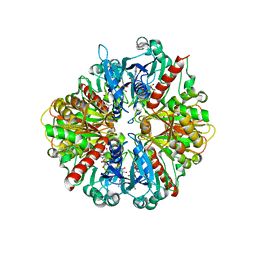 | | Structure of Glycosomal Glyceraldehyde-3-Phosphate Dehydrogenase from Trypanosoma cruzi in complex with the irreversible iodoacetate inhibitor | | 分子名称: | GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase, glycosomal, ... | | 著者 | Guido, R.V.C, Balliano, T.L, Andricopulo, A.D, Oliva, G. | | 登録日 | 2008-07-01 | | 公開日 | 2008-10-21 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Kinetic and Crystallographic Studies on Glyceraldehyde-3-Phosphate Dehydrogenase from Trypanosoma cruzi in Complex with Iodoacetate.
Letters in drug design & discovery, 6, 2009
|
|
3K2Y
 
 | | Crystal structure of protein lp_0118 from Lactobacillus plantarum,northeast structural genomics consortium target LpR91B | | 分子名称: | uncharacterized protein lp_0118 | | 著者 | Seetharaman, J, Lew, S, Vorobiev, S.M, Wang, D, Janjua, H, Cunningham, K, Owens, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2009-09-30 | | 公開日 | 2009-11-17 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of protein lp_0118 from Lactobacillus plantarum,northeast structural genomics consortium target LpR91B
To be Published
|
|
3JZ4
 
 | | Crystal structure of E. coli NADP dependent enzyme | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Succinate-semialdehyde dehydrogenase [NADP+] | | 著者 | Langendorf, C.G, Key, T.L.G, Fenalti, G, Kan, W.T, Buckle, A.M, Caradoc-Davies, T, Tuck, K.L, Law, R.H.P, Whisstock, J.C. | | 登録日 | 2009-09-22 | | 公開日 | 2010-03-16 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The X-ray crystal structure of Escherichia coli succinic semialdehyde dehydrogenase; structural insights into NADP+/enzyme interactions.
Plos One, 5, 2010
|
|
6CSJ
 
 | |
