1DG9
 
 | | CRYSTAL STRUCTURE OF BOVINE LOW MOLECULAR WEIGHT PTPASE COMPLEXED WITH HEPES | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, TYROSINE PHOSPHATASE | | 著者 | Zhang, M, Zhou, M, Van Etten, R.L, Stauffacher, C.V. | | 登録日 | 1999-11-23 | | 公開日 | 1999-12-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of bovine low molecular weight phosphotyrosyl phosphatase complexed with the transition state analog vanadate.
Biochemistry, 36, 1997
|
|
3QWO
 
 | |
258L
 
 | | AN ADAPTABLE METAL-BINDING SITE ENGINEERED INTO T4 LYSOZYME | | 分子名称: | CHLORIDE ION, LYSOZYME, ZINC ION | | 著者 | Wray, J.W, Baase, W.A, Ostheimer, G.J, Matthews, B.W. | | 登録日 | 1999-01-05 | | 公開日 | 2000-09-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Use of a non-rigid region in T4 lysozyme to design an adaptable metal-binding site.
Protein Eng., 13, 2000
|
|
1IAC
 
 | | REFINED 1.8 ANGSTROMS X-RAY CRYSTAL STRUCTURE OF ASTACIN, A ZINC-ENDOPEPTIDASE FROM THE CRAYFISH ASTACUS ASTACUS L. STRUCTURE DETERMINATION, REFINEMENT, MOLECULAR STRUCTURE AND COMPARISON WITH THERMOLYSIN | | 分子名称: | ASTACIN, MERCURY (II) ION | | 著者 | Gomis-Rueth, F.-X, Stoecker, W, Bode, W. | | 登録日 | 1994-05-09 | | 公開日 | 1994-08-31 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Refined 1.8 A X-ray crystal structure of astacin, a zinc-endopeptidase from the crayfish Astacus astacus L. Structure determination, refinement, molecular structure and comparison with thermolysin.
J.Mol.Biol., 229, 1993
|
|
4G7F
 
 | |
1ICE
 
 | | STRUCTURE AND MECHANISM OF INTERLEUKIN-1BETA CONVERTING ENZYME | | 分子名称: | INTERLEUKIN-1 BETA CONVERTING ENZYME, TETRAPEPTIDE ALDEHYDE | | 著者 | Wilson, K.P, Griffith, J.P, Kim, E.E, Navia, M.A. | | 登録日 | 1994-09-29 | | 公開日 | 1995-07-28 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure and mechanism of interleukin-1 beta converting enzyme.
Nature, 370, 1994
|
|
2JN5
 
 | |
3AOK
 
 | |
3R2Y
 
 | | MK2 kinase bound to Compound 1 | | 分子名称: | 2-(2-QUINOLIN-3-YLPYRIDIN-4-YL)-1,5,6,7-TETRAHYDRO-4H-PYRROLO[3,2-C]PYRIDIN-4-ONE, MALONATE ION, MAP kinase-activated protein kinase 2 | | 著者 | Oubrie, A, Leonard, P. | | 登録日 | 2011-03-15 | | 公開日 | 2011-05-25 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure-based lead identification of ATP-competitive MK2 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3AOX
 
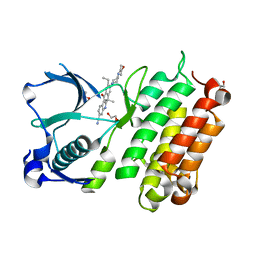 | | X-ray crystal structure of human anaplastic lymphoma kinase in complex with CH5424802 | | 分子名称: | 1,2-ETHANEDIOL, 9-ethyl-6,6-dimethyl-8-[4-(morpholin-4-yl)piperidin-1-yl]-11-oxo-6,11-dihydro-5H-benzo[b]carbazole-3-carbonitrile, ALK tyrosine kinase receptor | | 著者 | Nagel, S, Moertl, M, Jestel, A, Fukami, T.A. | | 登録日 | 2010-10-08 | | 公開日 | 2011-05-25 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | CH5424802, a selective ALK inhibitor capable of blocking the resistant gatekeeper mutant
Cancer Cell, 19, 2011
|
|
4FTD
 
 | |
2JMR
 
 | | NMR structure of the E. coli type 1 pilus subunit FimF | | 分子名称: | fimF | | 著者 | Gossert, A.D, Bettendorff, P, Puorger, C, Vetsch, M, Herrmann, T, Fiorito, F, Hiller, S, Glockshuber, R, Wuthrich, K. | | 登録日 | 2006-11-29 | | 公開日 | 2007-10-30 | | 最終更新日 | 2024-11-06 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of the Escherichia coli type 1 pilus subunit FimF and its interactions with other pilus subunits.
J.Mol.Biol., 375, 2008
|
|
5SRY
 
 | |
4G1B
 
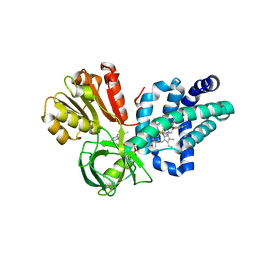 | | X-ray structure of yeast flavohemoglobin in complex with econazole | | 分子名称: | 1-[(2S)-2-[(4-CHLOROBENZYL)OXY]-2-(2,4-DICHLOROPHENYL)ETHYL]-1H-IMIDAZOLE, FLAVIN-ADENINE DINUCLEOTIDE, Flavohemoglobin, ... | | 著者 | El Hammi, E, Warkentin, E, Demmer, U, Baciou, L, Ermler, U. | | 登録日 | 2012-07-10 | | 公開日 | 2012-11-14 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Active site analysis of yeast flavohemoglobin based on its structure with a small ligand or econazole.
Febs J., 279, 2012
|
|
1R2R
 
 | | CRYSTAL STRUCTURE OF RABBIT MUSCLE TRIOSEPHOSPHATE ISOMERASE | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | 著者 | Aparicio, R, Ferreira, S.T, Polikarpov, I. | | 登録日 | 2003-09-29 | | 公開日 | 2003-12-23 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Closed conformation of the active site loop of rabbit muscle triosephosphate isomerase in the absence of substrate: evidence of conformational heterogeneity.
J.Mol.Biol., 334, 2003
|
|
3R0X
 
 | | Crystal structure of Selenomethionine incorporated apo D-serine deaminase from Salmonella tyhimurium | | 分子名称: | 1,2-ETHANEDIOL, D-serine dehydratase, SODIUM ION, ... | | 著者 | Bharath, S.R, Shveta, B, Savithri, H.S, Murthy, M.R.N. | | 登録日 | 2011-03-09 | | 公開日 | 2011-06-29 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Crystal structures of open and closed forms of D-serine deaminase from Salmonella typhimurium - implications on substrate specificity and catalysis
Febs J., 2011
|
|
5SOW
 
 | |
5SPN
 
 | |
5SET
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1(cc(nn1C)c2ccccn2)NC(=O)c3c(ccc(n3)C4CC4)Nc5cncnc5, micromolar IC50=>1.66968 | | 分子名称: | 6-cyclopropyl-N-[1-methyl-3-(pyridin-2-yl)-1H-pyrazol-5-yl]-3-[(pyrimidin-5-yl)amino]pyridine-2-carboxamide, MAGNESIUM ION, ZINC ION, ... | | 著者 | Joseph, C, Groebke-Zbinden, K, Benz, J, Schlatter, D, Rudolph, M.G. | | 登録日 | 2022-01-21 | | 公開日 | 2022-10-12 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
4KTN
 
 | | Dna gyrase atp binding domain of enterococcus faecalis in complex with a small molecule inhibitor ((3S)-1-[2-(PYRIDO[2,3-B]PYRAZIN-7-YLSULFANYL)-9H-PYRIMIDO[4,5-B]INDOL-4-YL]PYRROLIDIN-3-AMINE) | | 分子名称: | (3S)-1-[2-(pyrido[2,3-b]pyrazin-7-ylsulfanyl)-9H-pyrimido[4,5-b]indol-4-yl]pyrrolidin-3-amine, DNA gyrase subunit B | | 著者 | Bensen, D.C, Akers-rodriguez, S, Tari, L.W. | | 登録日 | 2013-05-20 | | 公開日 | 2014-01-15 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | A new class of type iia topoisomerase inhibitors with
broad-spectrum antibacterial activity
To be Published
|
|
2JII
 
 | | Structure of vaccinia related kinase 3 | | 分子名称: | 1,2-ETHANEDIOL, SERINE/THREONINE-PROTEIN KINASE VRK3 MOLECULE: VACCINIA RELATED KINASE 3 | | 著者 | Bunkoczi, G, Eswaran, J, Pike, A.C.W, Uppenberg, J, Ugochukwu, E, von Delft, F, Cooper, C, Salah, E, Savitsky, P, Burgess-Brown, N, Keates, T, Fedorov, O, Sobott, F, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Weigelt, J, Knapp, S. | | 登録日 | 2007-06-28 | | 公開日 | 2007-07-10 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of the pseudokinase VRK3 reveals a degraded catalytic site, a highly conserved kinase fold, and a putative regulatory binding site.
Structure, 17, 2009
|
|
259L
 
 | | AN ADAPTABLE METAL-BINDING SITE ENGINEERED INTO T4 LYSOZYME | | 分子名称: | CHLORIDE ION, COBALT (II) ION, PROTEIN (LYSOZYME) | | 著者 | Wray, J.W, Baase, W.A, Ostheimer, G.J, Matthews, B.W. | | 登録日 | 1999-02-10 | | 公開日 | 1999-04-12 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Use of a non-rigid region in T4 lysozyme to design an adaptable metal-binding site.
Protein Eng., 13, 2000
|
|
3V9M
 
 | |
2EP8
 
 | |
3ARA
 
 | | Discovery of Novel Uracil Derivatives as Potent Human dUTPase Inhibitors | | 分子名称: | 1-[3-({(2R)-2-[hydroxy(diphenyl)methyl]pyrrolidin-1-yl}sulfonyl)propyl]pyrimidine-2,4(1H,3H)-dione, Deoxyuridine 5'-triphosphate nucleotidohydrolase, MAGNESIUM ION | | 著者 | Chong, K.T, Miyakoshi, H, Miyahara, S, Fukuoka, M. | | 登録日 | 2010-11-25 | | 公開日 | 2010-12-08 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Synthesis and discovery of N-carbonylpyrrolidine- or N-sulfonylpyrrolidine-containing uracil derivatives as potent human deoxyuridine triphosphatase inhibitors
J.Med.Chem., 55, 2012
|
|
