3D2U
 
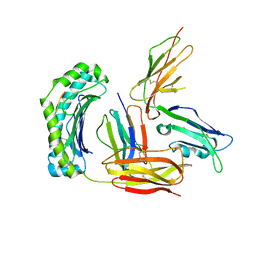 | | Structure of UL18, a Peptide-Binding Viral MHC Mimic, Bound to a Host Inhibitory Receptor | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Actin, Beta-2-microglobulin, ... | | 著者 | Yang, Z, Bjorkman, P.J. | | 登録日 | 2008-05-08 | | 公開日 | 2008-07-08 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Structure of UL18, a peptide-binding viral MHC mimic, bound to a host inhibitory receptor
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3GZI
 
 | |
3GY9
 
 | |
3H0N
 
 | |
3GZ7
 
 | |
3CLM
 
 | |
3CMQ
 
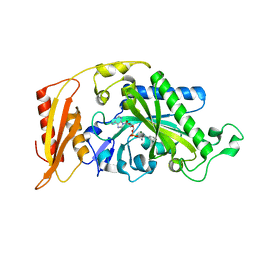 | | Crystal structure of human mitochondrial phenylalanine tRNA synthetase | | 分子名称: | ADENOSINE-5'-[PHENYLALANINYL-PHOSPHATE], MAGNESIUM ION, Phenylalanyl-tRNA synthetase, ... | | 著者 | Klipcan, L, Levin, I.L, Kessler, N, Moor, N, Finarov, I, Safro, M. | | 登録日 | 2008-03-24 | | 公開日 | 2008-07-01 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The tRNA-Induced Conformational Activation of Human Mitochondrial Phenylalanyl-tRNA Synthetase.
Structure, 16, 2008
|
|
3EUA
 
 | |
3E5D
 
 | |
3ECQ
 
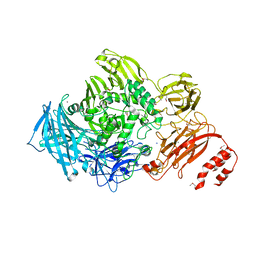 | | Endo-alpha-N-acetylgalactosaminidase from Streptococcus pneumoniae: SeMet structure | | 分子名称: | CALCIUM ION, Endo-alpha-N-acetylgalactosaminidase, GLYCEROL, ... | | 著者 | Caines, M.E.C, Zhu, H, Vuckovic, M, Strynadka, N.C.J. | | 登録日 | 2008-09-01 | | 公開日 | 2008-09-09 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | The structural basis for T-antigen hydrolysis by Streptococcus pneumoniae: a target for structure-based vaccine design.
J.Biol.Chem., 283, 2008
|
|
3ES4
 
 | |
3EQX
 
 | |
3ES1
 
 | |
3EUC
 
 | |
3ETN
 
 | |
3EU8
 
 | |
3GRD
 
 | |
3H3L
 
 | |
3H4Y
 
 | |
3GJU
 
 | |
3GZA
 
 | |
3GWP
 
 | |
3GZB
 
 | |
3GS9
 
 | |
3HBA
 
 | |
