1PHZ
 
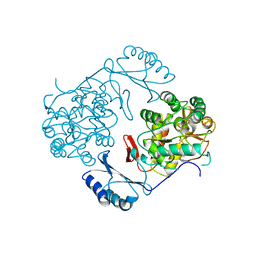 | | STRUCTURE OF PHOSPHORYLATED PHENYLALANINE HYDROXYLASE | | 分子名称: | FE (III) ION, PROTEIN (PHENYLALANINE HYDROXYLASE) | | 著者 | Kobe, B, Jennings, I.G, House, C.M, Michell, B.J, Cotton, R.G, Kemp, B.E. | | 登録日 | 1998-11-11 | | 公開日 | 1999-04-30 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis of autoregulation of phenylalanine hydroxylase.
Nat.Struct.Biol., 6, 1999
|
|
7L3H
 
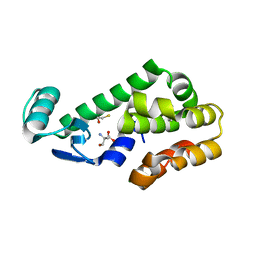 | | T4 Lysozyme L99A - ethylbenzene - RT | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Fischer, M, Bradford, S.Y.C. | | 登録日 | 2020-12-17 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L3B
 
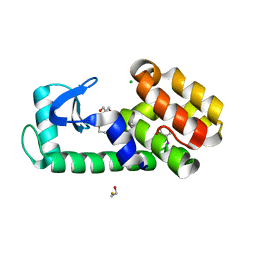 | | T4 Lysozyme L99A - iodobenzene - RT | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Fischer, M, Bradford, S.Y.C. | | 登録日 | 2020-12-17 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.27 Å) | | 主引用文献 | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L3G
 
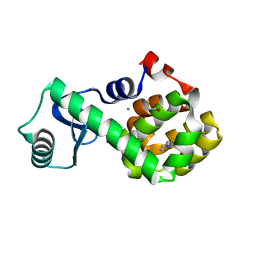 | | T4 Lysozyme L99A - 4-iodotoluene - cryo | | 分子名称: | 1-iodo-4-methylbenzene, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | 著者 | Fischer, M, Bradford, S.Y.C. | | 登録日 | 2020-12-17 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.27 Å) | | 主引用文献 | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
8U4E
 
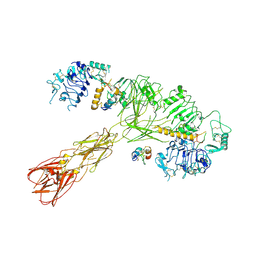 | | Cryo-EM structure of long form insulin receptor (IR-B) with three IGF2 bound, asymmetric conformation. | | 分子名称: | Insulin receptor, Insulin-like growth factor II | | 著者 | An, W, Hall, C, Li, J, Huang, A, Wu, J, Park, J, Bai, X.C, Choi, E. | | 登録日 | 2023-09-10 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Activation of the insulin receptor by insulin-like growth factor 2.
Nat Commun, 15, 2024
|
|
4MU0
 
 | | The structure of wt A. thaliana IGPD2 in complex with Mn2+ and 1,2,4-triazole at 1.3 A resolution | | 分子名称: | 1,2,4-TRIAZOLE, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | 著者 | Bisson, C, Britton, K.L, Sedelnikova, S.E, Baker, P.J, Rice, D.W. | | 登録日 | 2013-09-20 | | 公開日 | 2014-09-24 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Crystal Structures Reveal that the Reaction Mechanism of Imidazoleglycerol-Phosphate Dehydratase Is Controlled by Switching Mn(II) Coordination.
Structure, 23, 2015
|
|
2YIT
 
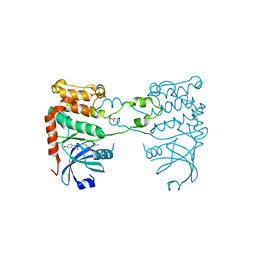 | | Structural analysis of checkpoint kinase 2 in complex with PV1162, a novel inhibitor | | 分子名称: | N-{4-[(1E)-N-carbamimidoylbutanehydrazonoyl]phenyl}-5-methoxy-1H-indole-2-carboxamide, NITRATE ION, SERINE/THREONINE-PROTEIN KINASE CHK2 | | 著者 | Lountos, G.T, Jobson, A.G, Tropea, J.E, Self, C, Zhang, G, Pommier, Y, Shoemaker, R.H, Waugh, D.S. | | 登録日 | 2011-05-16 | | 公開日 | 2011-09-07 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | X-Ray Structures of Checkpoint Kinase 2 in Complex with Inhibitors that Target its Gatekeeper-Dependent Hydrophobic Pocket.
FEBS Lett., 585, 2011
|
|
2W1L
 
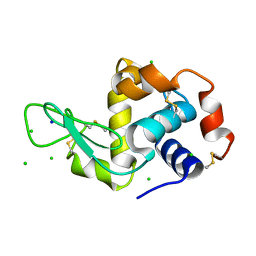 | | THE INTERDEPENDENCE OF WAVELENGTH, REDUNDANCY AND DOSE IN SULFUR SAD EXPERIMENTS: 0.979 a wavelength 991 images data | | 分子名称: | CHLORIDE ION, LYSOZYME C, SODIUM ION | | 著者 | Cianci, M, Helliwell, J.R, Suzuki, A. | | 登録日 | 2008-10-17 | | 公開日 | 2008-10-28 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | The Interdependence of Wavelength, Redundancy and Dose in Sulfur Sad Experiments.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
7L3F
 
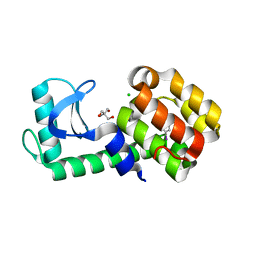 | | T4 Lysozyme L99A - 4-iodotoluene - RT | | 分子名称: | 1-iodo-4-methylbenzene, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | 著者 | Fischer, M, Bradford, S.Y.C. | | 登録日 | 2020-12-17 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
2C55
 
 | | Solution Structure of the Human Immunodeficiency Virus Type 1 p6 Protein | | 分子名称: | PROTEIN P6 | | 著者 | Fossen, T, Wray, V, Bruns, K, Rachmat, J, Henklein, P, Tessmer, U, Maczurek, A, Klinger, P, Schubert, U. | | 登録日 | 2005-10-25 | | 公開日 | 2005-11-02 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of the Human Immunodeficiency Virus Type 1 P6 Protein.
J.Biol.Chem., 280, 2005
|
|
1PER
 
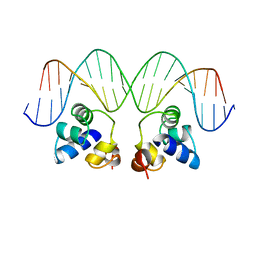 | |
5COX
 
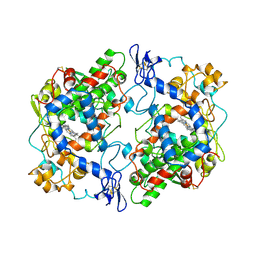 | |
4MUI
 
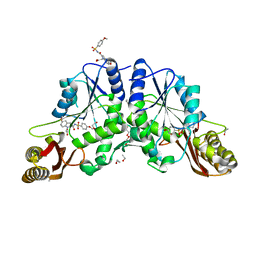 | |
4H6C
 
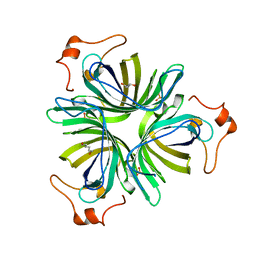 | |
7Q6I
 
 | | Vibrio maritimus FtsA 1-396 ATP and FtsN 1-29, bent tetramers in double filament arrangement | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell division protein FtsA, Cell division protein FtsN (polyAla model), ... | | 著者 | Nierhaus, T, Kureisaite-Ciziene, D, Lowe, J. | | 登録日 | 2021-11-07 | | 公開日 | 2022-09-21 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Bacterial divisome protein FtsA forms curved antiparallel double filaments when binding to FtsN.
Nat Microbiol, 7, 2022
|
|
7L3K
 
 | | T4 Lysozyme L99A - benzylacetate - cryo | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Fischer, M, Bradford, S.Y.C. | | 登録日 | 2020-12-17 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.11 Å) | | 主引用文献 | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
4N8X
 
 | |
8VAS
 
 | |
2W2G
 
 | | Human SARS coronavirus unique domain | | 分子名称: | NON-STRUCTURAL PROTEIN 3, SULFATE ION | | 著者 | Tan, J, Vonrhein, C, Smart, O.S, Bricogne, G, Bollati, M, Kusov, Y, Hansen, G, Mesters, J.R, Schmidt, C.L, Hilgenfeld, R. | | 登録日 | 2008-10-30 | | 公開日 | 2009-05-26 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | The Sars-Unique Domain (Sud) of Sars Coronavirus Contains Two Macrodomains that Bind G-Quadruplexes.
Plos Pathog., 5, 2009
|
|
7L3C
 
 | | T4 Lysozyme L99A - o-xylene - RT | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Fischer, M, Bradford, S.Y.C. | | 登録日 | 2020-12-17 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.31 Å) | | 主引用文献 | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7QV7
 
 | | Cryo-EM structure of Hydrogen-dependent CO2 reductase. | | 分子名称: | Hydrogen dependent carbon dioxide reductase subunit FdhF, Hydrogen dependent carbon dioxide reductase subunit HycB3, Hydrogen dependent carbon dioxide reductase subunit HycB4, ... | | 著者 | Dietrich, H.M, Righetto, R.D, Kumar, A, Wietrzynski, W, Schuller, S.K, Trischler, R, Wagner, J, Schwarz, F.M, Engel, B.D, Mueller, V, Schuller, J.M. | | 登録日 | 2022-01-19 | | 公開日 | 2022-07-06 | | 最終更新日 | 2022-08-10 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Membrane-anchored HDCR nanowires drive hydrogen-powered CO 2 fixation.
Nature, 607, 2022
|
|
4MV6
 
 | | Crystal Structure of Biotin Carboxylase from Haemophilus influenzae in Complex with Phosphonoacetamide | | 分子名称: | 1,2-ETHANEDIOL, Biotin carboxylase, PHOSPHONOACETAMIDE | | 著者 | Broussard, T.C, Pakhomova, S, Neau, D.B, Champion, T.S, Bonnot, R.J, Waldrop, G.L. | | 登録日 | 2013-09-23 | | 公開日 | 2015-01-14 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Structural Analysis of Substrate, Reaction Intermediate, and Product Binding in Haemophilus influenzae Biotin Carboxylase.
Biochemistry, 54, 2015
|
|
4NAO
 
 | | Crystal structure of EasH | | 分子名称: | 2-OXOGLUTARIC ACID, FE (II) ION, HEXAETHYLENE GLYCOL, ... | | 著者 | Janke, R, Havemann, J, Vogel, D, Keller, U, Loll, B. | | 登録日 | 2013-10-22 | | 公開日 | 2014-01-15 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.649 Å) | | 主引用文献 | Cyclolization of d-lysergic Acid alkaloid peptides.
Chem.Biol., 21, 2014
|
|
2YM0
 
 | | Truncated SipD from Salmonella typhimurium | | 分子名称: | CELL INVASION PROTEIN SIPD, GLYCEROL | | 著者 | Lunelli, M, Kolbe, M. | | 登録日 | 2011-06-06 | | 公開日 | 2011-08-17 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structure of PrgI-SipD: insight into a secretion competent state of the type three secretion system needle tip and its interaction with host ligands.
PLoS Pathog., 7, 2011
|
|
4NB0
 
 | | Crystal Structure of FosB from Staphylococcus aureus with BS-Cys9 disulfide at 1.62 Angstrom Resolution | | 分子名称: | CYSTEINE, GLYCEROL, Metallothiol transferase FosB, ... | | 著者 | Thompson, M.K, Goodman, M.C, Jagessar, K, Harp, J, Keithly, M.E, Cook, P.D, Armstrong, R.N. | | 登録日 | 2013-10-22 | | 公開日 | 2014-02-26 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Structure and Function of the Genomically Encoded Fosfomycin Resistance Enzyme, FosB, from Staphylococcus aureus.
Biochemistry, 53, 2014
|
|
