8OVG
 
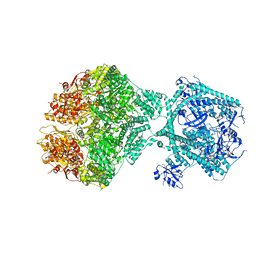 | | Human Mitochondrial Lon Y186E Mutant ADP Bound | | 分子名称: | Lon protease homolog, mitochondrial | | 著者 | Kereiche, S, Bauer, J.A, Matyas, P, Novacek, J, Kutejova, E. | | 登録日 | 2023-04-26 | | 公開日 | 2024-05-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (8.47 Å) | | 主引用文献 | Polyphosphate and tyrosine phosphorylation in the N-terminal domain of the human mitochondrial Lon protease disrupts its functions.
Sci Rep, 14, 2024
|
|
8OVF
 
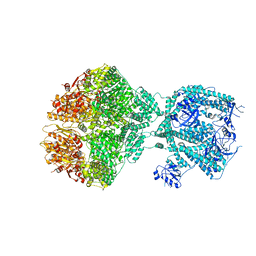 | | Human Mitochondrial Lon Y186F Mutant ADP Bound | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | 著者 | Kereiche, S, Bauer, J.A, Matyas, P, Novacek, J, Kutejova, E. | | 登録日 | 2023-04-26 | | 公開日 | 2024-05-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (7.23 Å) | | 主引用文献 | Polyphosphate and tyrosine phosphorylation in the N-terminal domain of the human mitochondrial Lon protease disrupts its functions.
Sci Rep, 14, 2024
|
|
8KG2
 
 | |
8OOI
 
 | | Full composite cryo-EM map of p97/VCP in ADP.Pi state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Cheng, T.C, Sakata, E, Schuetz, A.K. | | 登録日 | 2023-04-05 | | 公開日 | 2024-01-31 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (2.61 Å) | | 主引用文献 | Characterizing ATP processing by the AAA+ protein p97 at the atomic level.
Nat.Chem., 16, 2024
|
|
7FD4
 
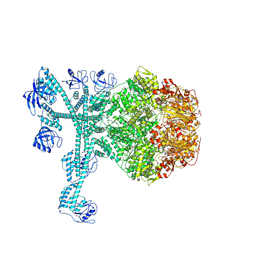 | | A complete three-dimensional structure of the Lon protease translocating a protein substrate (conformation 1) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Alpha-S1-casein, Lon protease, ... | | 著者 | Li, S, Hsieh, K, Kuo, C, Lee, S, Pintilie, G, Zhang, K, Chang, C. | | 登録日 | 2021-07-16 | | 公開日 | 2021-11-03 | | 実験手法 | ELECTRON MICROSCOPY (2.4 Å) | | 主引用文献 | Complete three-dimensional structures of the Lon protease translocating a protein substrate.
Sci Adv, 7, 2021
|
|
7FD5
 
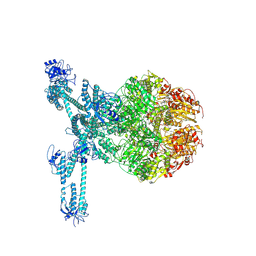 | | A complete three-dimensional structure of the Lon protease translocating a protein substrate (conformation 2) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Alpha-S1-casein, Lon protease, ... | | 著者 | Li, S, Hsieh, K, Kuo, C, Lee, S, Pintilie, G, Zhang, K, Chang, C. | | 登録日 | 2021-07-16 | | 公開日 | 2021-11-03 | | 実験手法 | ELECTRON MICROSCOPY (2.4 Å) | | 主引用文献 | Complete three-dimensional structures of the Lon protease translocating a protein substrate.
Sci Adv, 7, 2021
|
|
7CTG
 
 | | Human Origin Recognition Complex, ORC1-5 State I | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Origin recognition complex subunit 1, Origin recognition complex subunit 2, ... | | 著者 | Cheng, J, Li, N, Wang, X, Hu, J, Zhai, Y, Gao, N. | | 登録日 | 2020-08-18 | | 公開日 | 2021-01-06 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (5 Å) | | 主引用文献 | Structural insight into the assembly and conformational activation of human origin recognition complex.
Cell Discov, 6, 2020
|
|
7FIZ
 
 | | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex (conformation 3) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Lon protease, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Li, S, Hsieh, K, Kuo, C, Su, S, Huang, K, Zhang, K, Chang, C.I. | | 登録日 | 2021-08-01 | | 公開日 | 2021-11-24 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.28 Å) | | 主引用文献 | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex.
Sci Adv, 7, 2021
|
|
7FIE
 
 | | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex (conformation 2) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Lon protease, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Li, S, Hsieh, K, Kuo, C, Su, S, Huang, K, Zhang, K, Chang, C.I. | | 登録日 | 2021-07-31 | | 公開日 | 2021-11-24 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (2.36 Å) | | 主引用文献 | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex.
Sci Adv, 7, 2021
|
|
7FID
 
 | | Processive cleavage of substrate at individual proteolytic active sites of the Lon proteasecomplex (conformation 1) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Lon protease, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Li, S, Hsieh, K, Kuo, C, Su, S, Huang, K, Zhang, K, Chang, C.I. | | 登録日 | 2021-07-31 | | 公開日 | 2021-11-24 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (2.44 Å) | | 主引用文献 | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex.
Sci Adv, 7, 2021
|
|
7D63
 
 | | Cryo-EM structure of 90S preribosome with inactive Utp24 (state C) | | 分子名称: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | 著者 | Du, Y, Zhang, J, An, W, Ye, K. | | 登録日 | 2020-09-29 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (12.3 Å) | | 主引用文献 | Cryo-EM structure of 90S preribosome with inactive Utp24 (state C)
To Be Published
|
|
7D5T
 
 | | Cryo-EM structure of 90S preribosome with inactive Utp24 (state F1) | | 分子名称: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | 著者 | Du, Y, Zhang, J, An, W, Ye, K. | | 登録日 | 2020-09-28 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (6 Å) | | 主引用文献 | Cryo-EM structure of 90S preribosome with inactive Utp24 (state F1)
To Be Published
|
|
7D5S
 
 | | Cryo-EM structure of 90S preribosome with inactive Utp24 (state A2) | | 分子名称: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S12, ... | | 著者 | Du, Y, Zhang, J, An, W, Ye, K. | | 登録日 | 2020-09-28 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Cryo-EM structure of 90S preribosome with inactive Utp24 (state A2)
To Be Published
|
|
7D4I
 
 | | Cryo-EM structure of 90S small ribosomal precursors complex with the DEAH-box RNA helicase Dhr1 (State F) | | 分子名称: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | 著者 | Du, Y, Zhang, J, An, W, Ye, K. | | 登録日 | 2020-09-24 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Cryo-EM structure of 90S small ribosomal precursors complex with Dhr1
To Be Published
|
|
6AMN
 
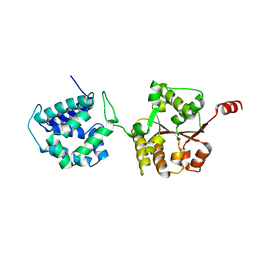 | | Crystal Structure of Hsp104 N Domain | | 分子名称: | Heat shock protein 104 | | 著者 | Lee, S. | | 登録日 | 2017-08-10 | | 公開日 | 2017-11-29 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.816 Å) | | 主引用文献 | Overlapping and Specific Functions of the Hsp104 N Domain Define Its Role in Protein Disaggregation.
Sci Rep, 7, 2017
|
|
6AHF
 
 | | CryoEM Reconstruction of Hsp104 N728A Hexamer | | 分子名称: | Heat shock protein 104, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | 著者 | Zhang, X, Zhang, L, Zhang, S. | | 登録日 | 2018-08-17 | | 公開日 | 2019-02-13 | | 最終更新日 | 2019-04-10 | | 実験手法 | ELECTRON MICROSCOPY (6.78 Å) | | 主引用文献 | Heat shock protein 104 (HSP104) chaperones soluble Tau via a mechanism distinct from its disaggregase activity.
J. Biol. Chem., 294, 2019
|
|
6AP1
 
 | | Vps4p-Vta1p complex with peptide binding to the central pore of Vps4p | | 分子名称: | ACE-ASP-GLU-ILE-VAL-ASN-LYS-VAL-LEU-NH2, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | 著者 | Han, H, Monroe, N, Shen, P, Sundquist, W.I, Hill, C.P. | | 登録日 | 2017-08-16 | | 公開日 | 2017-12-06 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | The AAA ATPase Vps4 binds ESCRT-III substrates through a repeating array of dipeptide-binding pockets.
Elife, 6, 2017
|
|
6AZY
 
 | | Crystal structure of Hsp104 R328M/R757M mutant from Calcarisporiella thermophila | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Heat shock protein Hsp104 | | 著者 | Michalska, K, Bigelow, L, Hatzos-Skintges, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2017-09-13 | | 公開日 | 2018-10-03 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure of Calcarisporiella thermophila Hsp104 Disaggregase that Antagonizes Diverse Proteotoxic Misfolding Events.
Structure, 27, 2019
|
|
6B5D
 
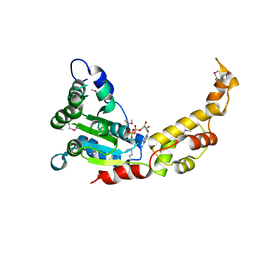 | |
6AZ0
 
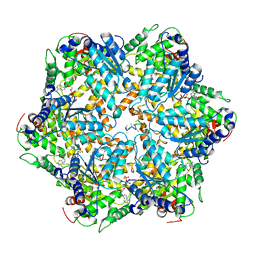 | | Mitochondrial ATPase Protease YME1 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Puchades, C, Rampello, A.J, Shin, M, Giuliano, C, Wiseman, R.L, Glynn, S.E, Lander, G.C. | | 登録日 | 2017-09-09 | | 公開日 | 2017-11-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure of the mitochondrial inner membrane AAA+ protease YME1 gives insight into substrate processing.
Science, 358, 2017
|
|
6B5C
 
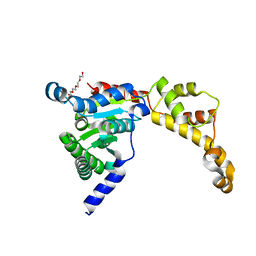 | |
6BLB
 
 | | 1.88 Angstrom Resolution Crystal Structure Holliday Junction ATP-dependent DNA Helicase (RuvB) from Pseudomonas aeruginosa in Complex with ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvB, TRIETHYLENE GLYCOL | | 著者 | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-11-09 | | 公開日 | 2017-11-22 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | 1.88 Angstrom Resolution Crystal Structure Holliday Junction ATP-dependent DNA Helicase (RuvB) from Pseudomonas aeruginosa in Complex with ADP.
To be Published
|
|
6BMF
 
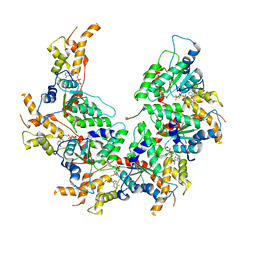 | | Vps4p-Vta1p complex with peptide binding to the central pore of Vps4p | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | 著者 | Han, H, Monroe, N, Shen, P, Sundquist, W.I, Hill, C.P. | | 登録日 | 2017-11-14 | | 公開日 | 2017-12-06 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | The AAA ATPase Vps4 binds ESCRT-III substrates through a repeating array of dipeptide-binding pockets.
Elife, 6, 2017
|
|
6CHS
 
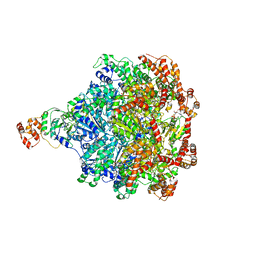 | | Cdc48-Npl4 complex in the presence of ATP-gamma-S | | 分子名称: | MAGNESIUM ION, Npl4, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Kim, K.H, Bodnar, N.O, Walz, T, Rapoport, T.A. | | 登録日 | 2018-02-22 | | 公開日 | 2018-07-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structure of the Cdc48 ATPase with its ubiquitin-binding cofactor Ufd1-Npl4.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6D00
 
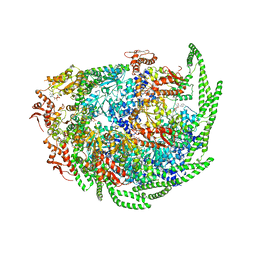 | | Calcarisporiella thermophila Hsp104 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Calcarisporiella thermophila Hsp104 | | 著者 | Zhang, K, Pintilie, G. | | 登録日 | 2018-04-09 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structure of Calcarisporiella thermophila Hsp104 Disaggregase that Antagonizes Diverse Proteotoxic Misfolding Events.
Structure, 27, 2019
|
|
