1IY0
 
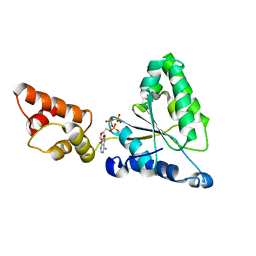 | | Crystal structure of the FtsH ATPase domain with AMP-PNP from Thermus thermophilus | | 分子名称: | ATP-dependent metalloprotease FtsH, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Niwa, H, Tsuchiya, D, Makyio, H, Yoshida, M, Morikawa, K. | | 登録日 | 2002-07-10 | | 公開日 | 2002-11-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Hexameric ring structure of the ATPase domain of the membrane-integrated metalloprotease FtsH from Thermus thermophilus HB8
Structure, 10, 2002
|
|
3J97
 
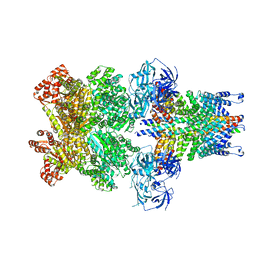 | | Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State II) | | 分子名称: | Alpha-soluble NSF attachment protein, Synaptosomal-associated protein 25, Syntaxin-1A, ... | | 著者 | Zhao, M, Wu, S, Cheng, Y, Brunger, A.T. | | 登録日 | 2014-12-05 | | 公開日 | 2015-01-28 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (7.8 Å) | | 主引用文献 | Mechanistic insights into the recycling machine of the SNARE complex.
Nature, 518, 2015
|
|
3HU2
 
 | | Structure of p97 N-D1 R86A mutant in complex with ATPgS | | 分子名称: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | 著者 | Tang, W.-K. | | 登録日 | 2009-06-12 | | 公開日 | 2010-06-16 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | A novel ATP-dependent conformation in p97 N-D1 fragment revealed by crystal structures of disease-related mutants.
Embo J., 29, 2010
|
|
3KDS
 
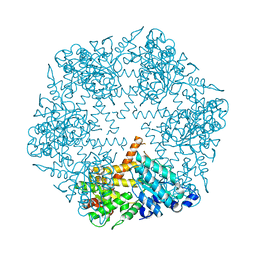 | | apo-FtsH crystal structure | | 分子名称: | Cell division protein FtsH, N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-3-naphthalen-2-yl-L-alanyl-L-alaninamide, ZINC ION | | 著者 | Bieniossek, C, Niederhauser, B, Baumann, U. | | 登録日 | 2009-10-23 | | 公開日 | 2009-12-01 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.601 Å) | | 主引用文献 | The crystal structure of apo-FtsH reveals domain movements necessary for substrate unfolding and translocation
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1LV7
 
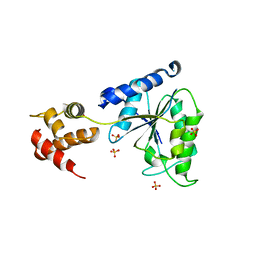 | | Crystal Structure of the AAA domain of FtsH | | 分子名称: | FtsH, SULFATE ION | | 著者 | Krzywda, S, Brzozowski, A.M, Verma, C, Karata, K, Ogura, T, Wilkinson, A.J. | | 登録日 | 2002-05-26 | | 公開日 | 2002-10-09 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The crystal structure of the AAA domain of the ATP-dependent protease FtsH of Escherichia coli at 1.5 A resolution.
Structure, 10, 2002
|
|
1IN7
 
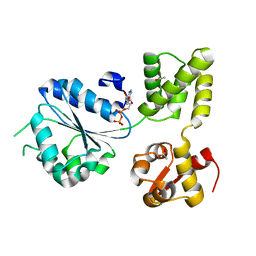 | | THERMOTOGA MARITIMA RUVB R170A | | 分子名称: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, HOLLIDAY JUNCTION DNA HELICASE RUVB | | 著者 | Putnam, C.D, Clancy, S.B, Tsuruta, H, Wetmur, J.G, Tainer, J.A. | | 登録日 | 2001-05-12 | | 公開日 | 2001-08-08 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure and mechanism of the RuvB Holliday junction branch migration motor.
J.Mol.Biol., 311, 2001
|
|
1IY2
 
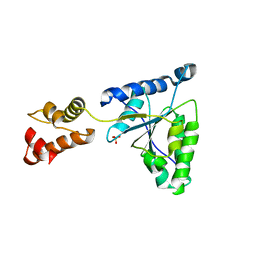 | | Crystal structure of the FtsH ATPase domain from Thermus thermophilus | | 分子名称: | ATP-dependent metalloprotease FtsH, SULFATE ION | | 著者 | Niwa, H, Tsuchiya, D, Makyio, H, Yoshida, M, Morikawa, K. | | 登録日 | 2002-07-10 | | 公開日 | 2002-11-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Hexameric ring structure of the ATPase domain of the membrane-integrated metalloprotease FtsH from Thermus thermophilus HB8
Structure, 10, 2002
|
|
1IN5
 
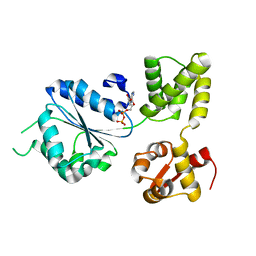 | | THERMOGOTA MARITIMA RUVB A156S MUTANT | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, HOLLIDAY JUNCTION DNA HELICASE RUVB | | 著者 | Putnam, C.D, Clancy, S.B, Tsuruta, H, Gonzalez, S, Wetmur, J.G, Tainer, J.A. | | 登録日 | 2001-05-12 | | 公開日 | 2001-08-08 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure and mechanism of the RuvB Holliday junction branch migration motor.
J.Mol.Biol., 311, 2001
|
|
8D6X
 
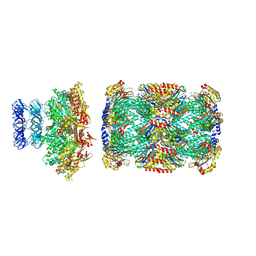 | |
8D6Y
 
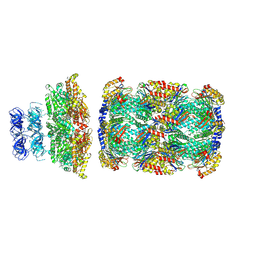 | |
6IP2
 
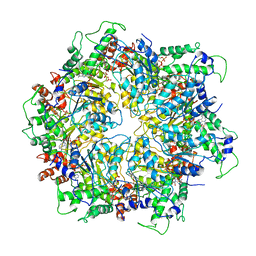 | | NSF-D1D2 part in the whole 20S complex | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Vesicle-fusing ATPase | | 著者 | Huang, X, Sun, S, Wang, X, Fan, F, Zhou, Q, Sui, S.F. | | 登録日 | 2018-11-01 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Mechanistic insights into the SNARE complex disassembly.
Sci Adv, 5, 2019
|
|
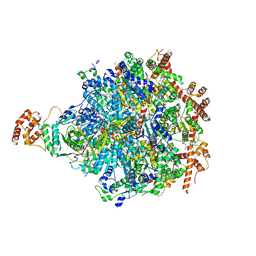
8DAR
 
 | |
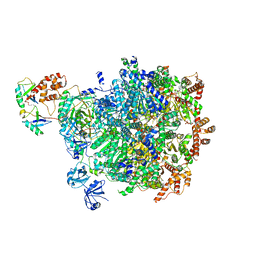
8DAU
 
 | |
8DAW
 
 | |
8DAS
 
 | |
8DAT
 
 | |
8DAV
 
 | |
6J2N
 
 | | yeast proteasome in substrate-processing state (C3-b) | | 分子名称: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | 著者 | Cong, Y. | | 登録日 | 2019-01-02 | | 公開日 | 2019-03-20 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (7.5 Å) | | 主引用文献 | Structural Snapshots of 26S Proteasome Reveal Tetraubiquitin-Induced Conformations.
Mol. Cell, 73, 2019
|
|
6J30
 
 | | yeast proteasome in Ub-engaged state (C2) | | 分子名称: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | 著者 | Cong, Y. | | 登録日 | 2019-01-03 | | 公開日 | 2019-03-20 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Structural Snapshots of 26S Proteasome Reveal Tetraubiquitin-Induced Conformations.
Mol. Cell, 73, 2019
|
|
6J2C
 
 | | Yeast proteasome in translocation competent state (C3-a) | | 分子名称: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | 著者 | Cong, Y. | | 登録日 | 2019-01-01 | | 公開日 | 2019-03-13 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (7 Å) | | 主引用文献 | Structural Snapshots of 26S Proteasome Reveal Tetraubiquitin-Induced Conformations.
Mol. Cell, 73, 2019
|
|
6J2X
 
 | | Yeast proteasome in resting state (C1-a) | | 分子名称: | 26S PROTEASE REGULATORY SUBUNIT 4 HOMOLOG, 26S PROTEASOME REGULATORY SUBUNIT RPN5, 26S proteasome complex subunit SEM1, ... | | 著者 | Cong, Y. | | 登録日 | 2019-01-03 | | 公開日 | 2019-03-13 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural Snapshots of 26S Proteasome Reveal Tetraubiquitin-Induced Conformations.
Mol. Cell, 73, 2019
|
|
6JPU
 
 | |
6J2Q
 
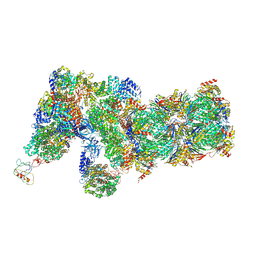 | | Yeast proteasome in Ub-accepted state (C1-b) | | 分子名称: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | 著者 | Cong, Y. | | 登録日 | 2019-01-02 | | 公開日 | 2019-03-13 | | 最終更新日 | 2019-04-10 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural Snapshots of 26S Proteasome Reveal Tetraubiquitin-Induced Conformations.
Mol. Cell, 73, 2019
|
|
8DR3
 
 | |
8DR7
 
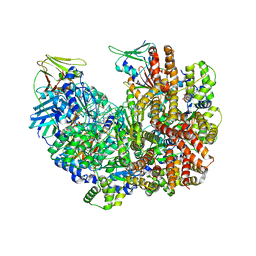 | | Open state of RFC:PCNA bound to a nicked dsDNA | | 分子名称: | DNA (26-MER), DNA (5'-D(P*AP*GP*GP*GP*GP*GP*GP*GP*GP*GP*G)-3'), DNA (5'-D(P*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), ... | | 著者 | Schrecker, M, Hite, R.K. | | 登録日 | 2022-07-20 | | 公開日 | 2022-08-17 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
